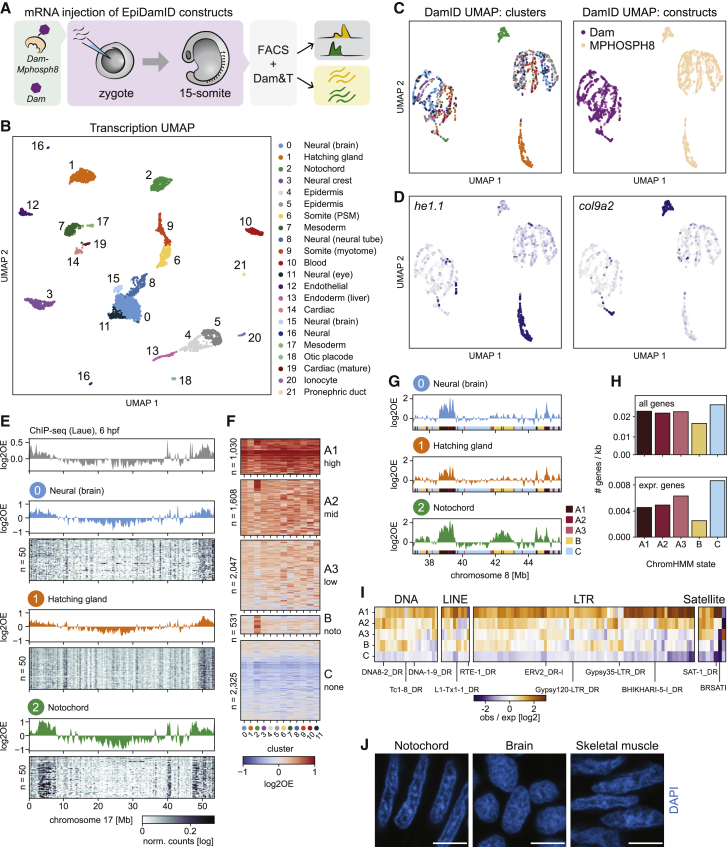
Figure 5.
Notochord-specific H3K9me3 enrichment in the zebrafish embryo
(A) Schematic representation of the experimental design and workflow.
(B) UMAP based on the transcriptional readout of all single-cell samples passing CEL-Seq2 thresholds (n = 3902).
(C) UMAP based on the genomic readout of all single-cell samples passing DamID thresholds (n = 2833). Samples are colored by transcriptional cluster (left) and Dam-targeting domain (right).
(D) Expression of the hatching gland marker he1.1 (left) and the notochord marker col9a2 (right) projected onto the DamID UMAP.
(E) Genomic H3K9me3 signal over chromosome 17. Top track: H3K9me3 ChIP-seq signal of 6-hpf embryo. Remaining tracks: combined single-cell Dam-MPHOSPH8 data for clusters 0–2. Heatmaps show the depth-normalized Dam-MPHOSPH8 data of the 50 richest cells.
(F) Heatmap showing the cluster-specific average H3K9me3 enrichment over all domains called per ChromHMM state. Per state, domains were clustered using hierarchical clustering.
(G) Genomic H3K9me3 signal over a part of chromosome 8 for clusters 0–2. The colored regions at the bottom of each track indicate the ChromHMM state.
(H) Gene density of all genes (top) and expressed genes (bottom) per state.
(I) Enrichment of repeats among the ChromHMM states. Example repeats are indicated.
(J) Representative images of DAPI staining in cryosections of zebrafish embryos at 15-somite stage. Scale bars represent 4 μm.
See also Figures S5 and S6.