Fig. 1.
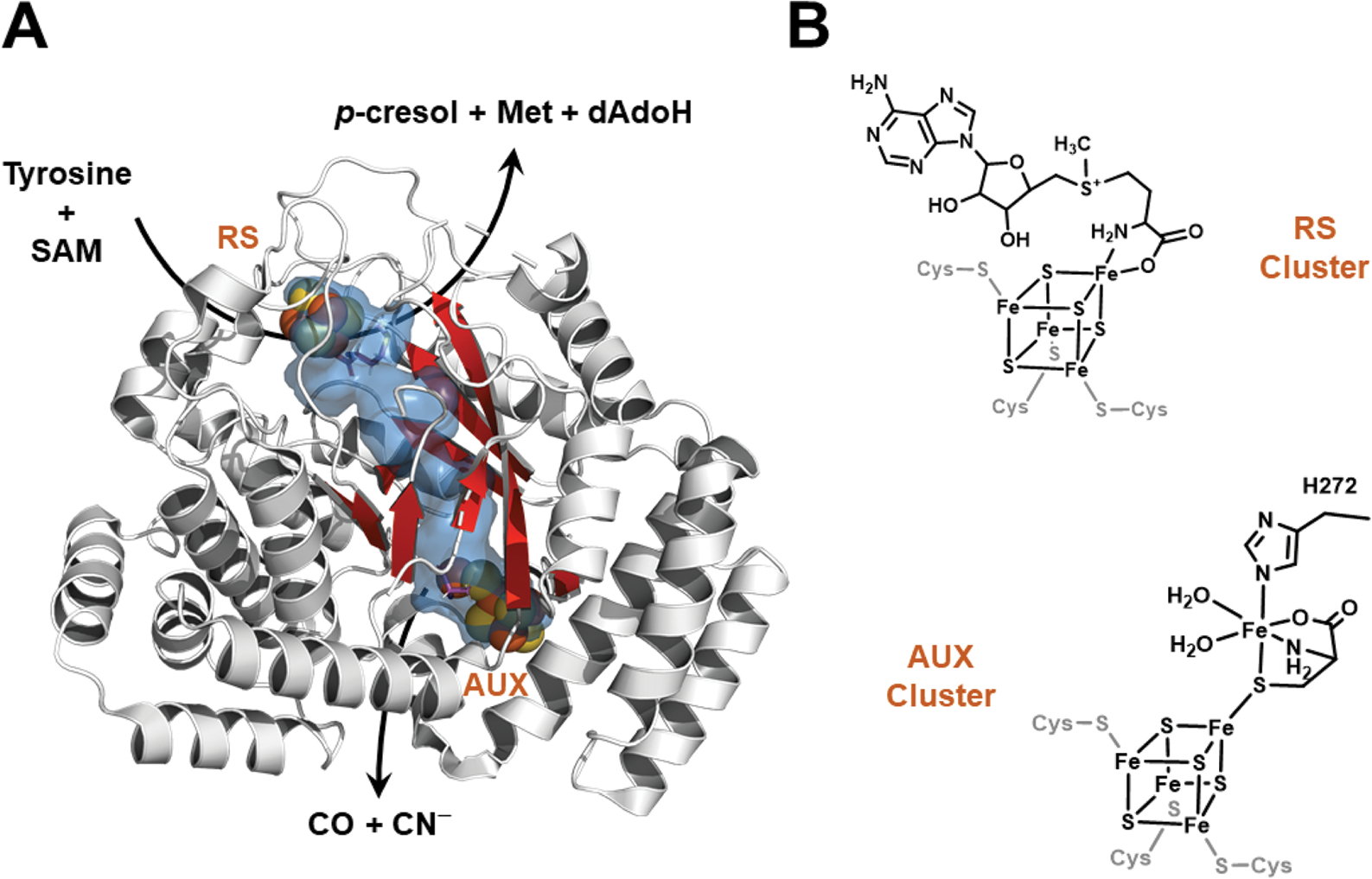
HydG structure (left) and iron–sulfur cluster states (right). A. The X-ray crystal structure of Thermoanaerobacter italicus HydG (4WCX.pdb). The radical SAM [4Fe−4S] cluster where tyrosine is cleaved to p-cresol and dehydroglycine (DHG) is at the top of the barrel and is denoted by “RS”, while the auxiliary cluster is located 24 Å away at the bottom of the barrel (denoted by “AUX”). The internal cavity calculation (shown in blue) was performed with Pymol (PyMOL Molecular Graphics System, Version 2.2.3 Schrödinger, LLC) using the Caver3 plugin; this internal tunnel connects solvent (near the RS cluster) to the AUX cluster, and is the proposed pathway by which DHG migrates from the RS to the AUX clusters. Residues 315–325 are not displayed because the helix they form hides the β-barrel, and thus precludes observation of the internal tunnel. B. Line drawings of the RS and AUX clusters of HydG. The conserved His272 residue that provides the only HydG-derived ligand to the dangler Fe is highlighted (amino acid numbering corresponds to Clostridium acetobutylicum).
