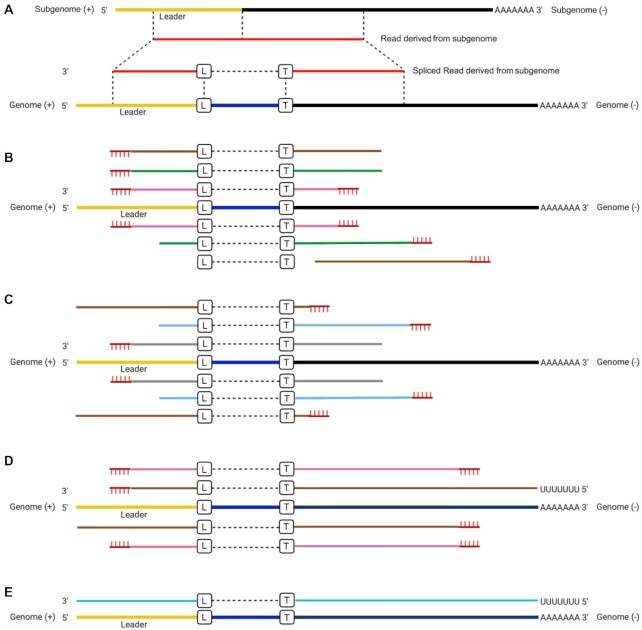
Figure 1.
(A) Illustration of reads derived from subgenomic mRNAs (sgmRNAs) mapped onto the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reference genome with a splicing method. We note that splicing does not occur in coronaviruses, but this is the apparent observation of a fusion event between different parts of the genome. (B, C) Illustration of the possible type of reads mapped on the SARS-CoV-2 reference genome for the paired-end Illumina amplicon sequencing, where the lines with the same color implied paired reads. (D) Nanopore amplicon sequencing and (E) Nanopore direct RNA sequencing of the SARS-CoV-2 genome and sgmRNAs. L and B in the boxes indicate the leader–transcriptional regulatory sequence (TRS) breaking sites on the leader side and TRS side, respectively, although we note these are where the apparent fusion site occurs. Yellow indicates the leader region, black is the TRS and gene sequence, and red indicates a sequence read that maps to the SARS-CoV-2 sequence. Blue is a sequence that is present between the leader sequence and the TRS. For (B) and (C), the same color (brown, green, and pink) indicates that same paired read. For (B), the paired read contains both primers. For (C), the gray and light blue color is a paired read but only contains one primer sequence at any end. The vertical hash lines on (B), (C), and (D) indicate the position of a primer.