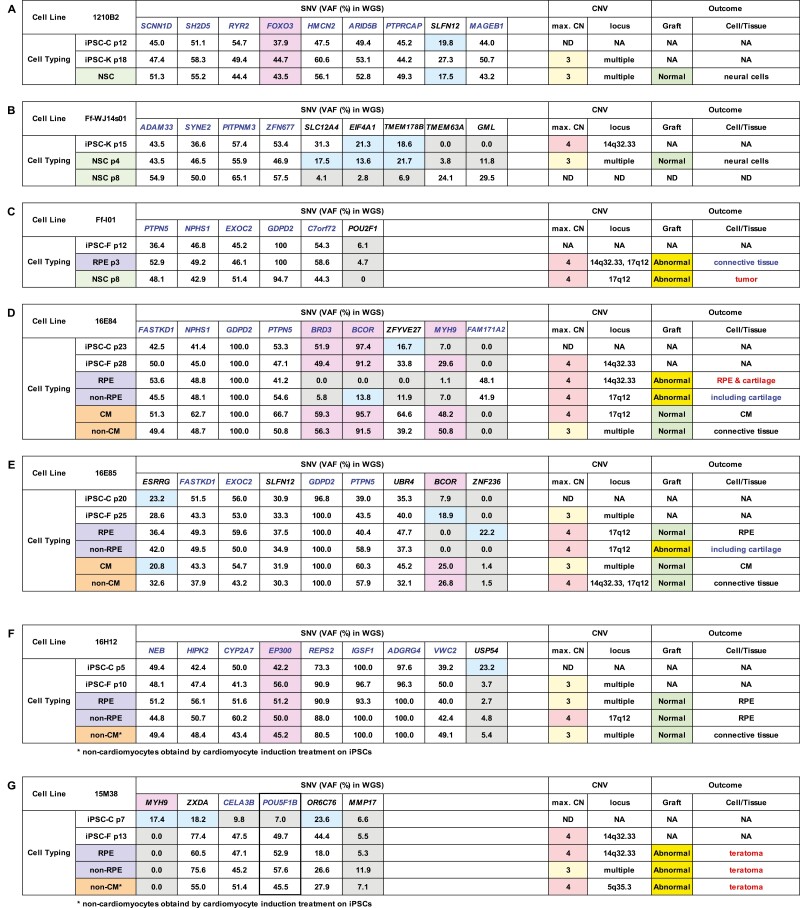
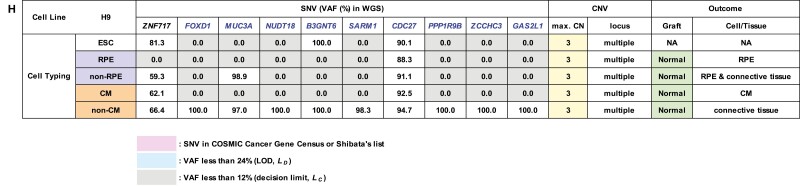
Table 1.
Genetic profiles of iPSC clones and their outcomes after transplantation into NOG mice.
VAF (%) of genes listed in the COSMIC ver.88 database determined by WGS and copy number variants of iPSC clones 1210B2 (A), Ff-WJ14s01 (B), Ff-I01 (C), 16E84 (D), 16E85 (E), 16H12 (F), or 15M38 (G) and their derivatives are shown in the table. Genetic mutations detected by WGS are SNVs, except deletions in BCOR in 16E84 (D) and 16E85 (E). VAFs less than 24% (below the detection limit [LOD]) are shown in blue cells, and VAFs less than 12% (below the decision limit) are shown in gray cells. VAFs of genes in the Census database and Shibata’s List showing values above the LOD are highlighted with a pinkish color. Gene whose VAFs reached around 50% or 100% related to the clonality of cells are shown in blue. VAFs of POU5F1B in 15M38 suggesting the integration of extrinsic POU5F1 (thick border). H9 ESCs and their derivatives (H) were used as controls. Maximum copy number (max.CN) of relevant cell clones and the loci showing CN = 4, if detected, are added to the table. Notable or abnormal findings of the transplants are described in blue or red in the table where applicable (Fig. 2). p: passage number, NA: not applicable, ND: not determined, NSC: neural stem cell, iPSC-C: iPSC cultured in CiRA, iPSC-K: iPSC transferred to Keio University used for differentiation and engrafting, iPSC-F: iPSC transferred to Foundation for Biomedical Research and Innovation (FBRI) used for differentiation and engrafting, RPE: retinal pigment epithelial cell, CM: cardiomyocyte. Many of the VAF values for the genes listed in the table reached nearly 50% in autosomal chromosomes and nearly 100% in the X chromosome if a male sample was used, suggesting that all of the iPSC clones tested in this study consisted of clonal expansion from a single cell and its derivatives.
Non-cardiomyocytes obtained by cardiomyocyte induction treatment on iPSCs.