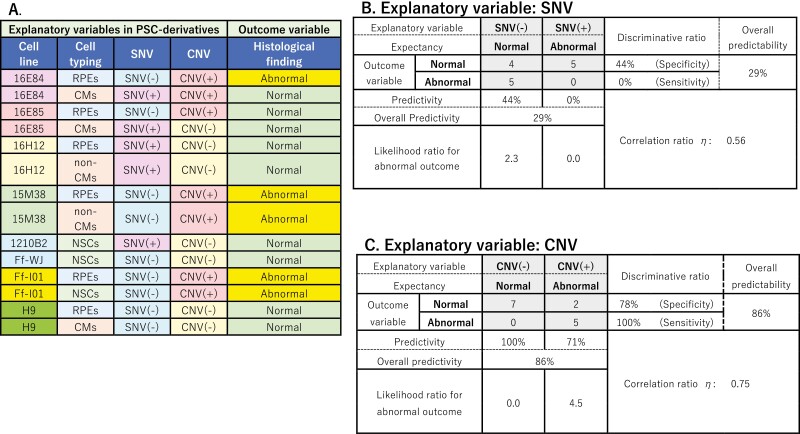
Table 2.
Outcome of the transplants of the cells with SNVs in the cancer-related genes or with copy number exceeding 3.
A. Table of iPSC-derivatives and their histology after transplantation. Cell typing: identities of differentiated cells transplanted, RPEs: retinal pigment epithelial cells, CMs: cardiomyocytes, NSCs: neural stem cells. All of the mutations in cancer-related genes listed in the table are SNVs, except deletions in BCORE in 84-CMs or16E85-CMs. SNV (+): cells with SNVs/del in cancer-related genes, SNV (−): cells with no detectable SNVs/del in cancer-related genes, CNV (+): cells with a maximum copy number exceeding 3, CNV (−): cells with copy numbers of 3 or below. B: Predictivity of abnormal tissue formation after transplantation by detecting cancer-related SNVs/del in the final product. C: Predictivity of abnormal tissue formation after transplantation by detecting a maximum copy number exceeding 3 in the final product. Categorical variables were analyzed with Hayashi’s quantification method type II.