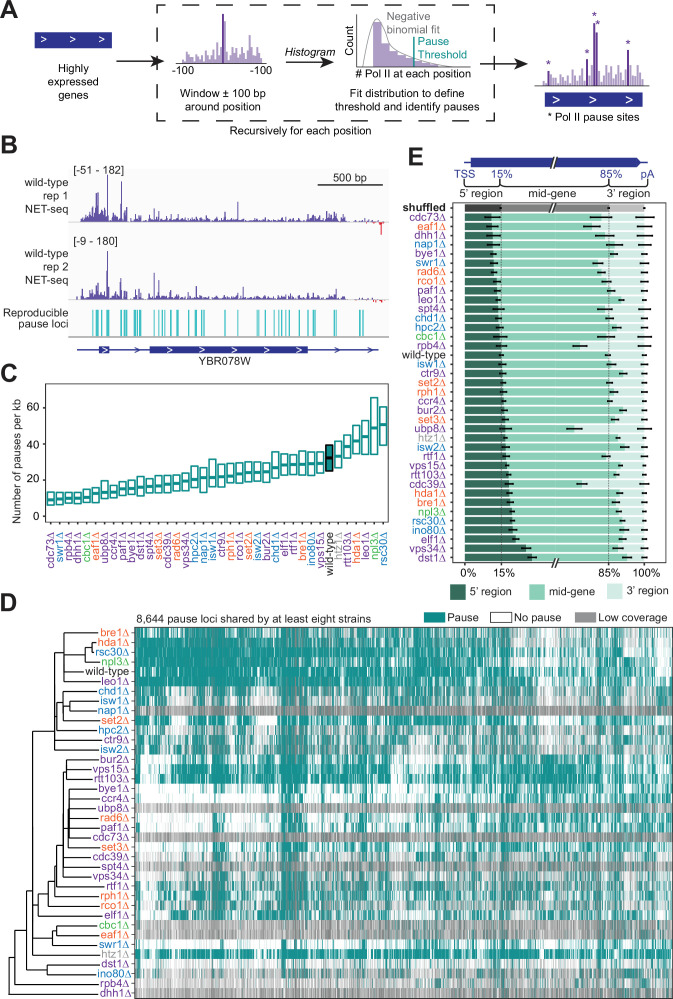
Figure 4. Trends in polymerase II (Pol II) pausing behavior at single-nucleotide resolution across deletion strains.
(A) Cartoon illustrating algorithm for robust and reproducible Pol II pause detection. (B) Example of Pol II density on the positive (purple) and negative (red) strands, as measured by native elongating transcript sequencing (NET-seq) in two wild-type replicates. Pauses that meet the 1% irreproducibility discovery rate (IDR) reproducibility threshold are shown as blue vertical lines. (C) Boxplot of the distribution of Pol II pause densities, the number of pauses per kilobase examined, in each deletion strain, ordered by median pausing density. Whiskers and outliers were removed for visualization. (D) Hierarchically clustered heatmap of 8644 Pol II pause loci across the genome reveals locations of pauses shared by multiple deletion strains. Heatmap is colored based on if that locus was identified as a pause (teal), not a pause (white), or if there was not sufficient coverage to determine pause status (gray). Analyses conducted only on deletion strains with biological replicates and only at loci at which there was enough coverage to determine the absence of a Pol II pause in at least one deletion strain. (E) The percent of Pol II pause loci located in the 5’ gene region, mid-gene, and 3’ gene region varies across deletion strains. The 5’ gene region was identified for each well-expressed gene as extending from the transcription start site to the 15th percentile of the gene length. Similarly, the 3’ gene region was defined as the last 15th percentile of the gene length, with the mid-gene region spanning in between. The control (gray) was created by scrambling all identified pauses across all deletion strains within the genes they were identified in. Rows are ordered by the percent of pauses found in the 5’ region. Bars represent the 95% confidence intervals across all expressed genes.