Abstract
Neuroblastoma is one of the most common pediatric cancers. This study used machine learning (ML) to predict the mortality and a few other investigated intermediate outcomes of neuroblastoma patients non-invasively from CT images. Performances of multiple ML algorithms over retrospective CT images of 65 neuroblastoma patients are analyzed. An artificial neural network (ANN) is used on tumor radiomic features extracted from 3D CT images. A pre-trained 2D convolutional neural network (CNN) is used on slices of the same images. ML models are trained for various pathologically investigated outcomes of these patients. A subspecialty-trained pediatric radiologist independently reviewed the manually segmented primary tumors. Pyradiomics library is used to extract 105 radiomic features. Six ML algorithms are compared to predict the following outcomes: mortality, presence or absence of metastases, neuroblastoma differentiation, mitosis-karyorrhexis index (MKI), presence or absence of MYCN gene amplification, and presence of image-defined risk factors (IDRF). The prediction ranges over multiple experiments are measured using the area under the receiver operating characteristic (ROC-AUC) for comparison. Our results show that the radiomics-based ANN method slightly outperforms the other algorithms in predicting all outcomes except classification of the grade of neuroblastic differentiation, for which the elastic regression model performed the best. Contributions of the article are twofold: (1) noninvasive models for the prognosis from CT images of neuroblastoma, and (2) comparison of relevant ML models on this medical imaging problem.
Keywords: Neuroblastoma, CT, Radiomics, Machine learning, Neural network
Introduction
Neuroblastoma is the most common extracranial malignant solid tumor in children and is responsible for approximately 15% of childhood cancer deaths [1]. It derives from adrenal glands or parasympathetic nerve tissue around the spine and presents as masses in the neck, chest, abdomen, pelvis, or paraspinal tissue. Neuroblastomas have a high degree of heterogeneity concerning both histology and clinical behavior [2, 3]. The clinical staging only based on the tumor encasement or invasion, such as the international neuroblastoma risk group (INRG) staging system, may overlook the tumor heterogeneity and may not be sufficient for prognosis assessment [4]. Some tumors of the same INRG stage may have a poor prognosis, while some other tumors have an excellent response to treatment or even the possibility of spontaneous regression and benign transformation [5, 6]. Therefore, assessment of multiple prognostic factors is critical to evaluate pretreatment risk stratification and allow early and effective treatment interventions for highly malignant tumors. In addition to the clinical staging, more risk factors have been found to be associated with prognosis [7]. For example, histologic indicators, grade of neuroblastic differentiation, and mitosis-karyorrhexis index (MKI) were found to have a prognostic value in neuroblastoma patients [8]. However, it is not easy to establish the relationship between the neuroblastoma tumor heterogeneity and those outcomes.
Radiomics is a promising way to assess the intra-tumoral heterogeneity through imaging and to assess prognosis [7]. Radiomics means a set of statistical features on an image. Instead of analyzing small tissue samples from biopsy, radiomic features are derived from the entire primary tumor volume. Thus, radiomics can provide important heterogeneity information of the entire tumor. Many studies of other cancers [9–11] have shown significant relationships between radiomics and different investigated variables such as hormone status, tumor proliferation, and gene expression patterns. For example, Jackson et al. [9] have shown that radiomics is highly correlated with overexpression of epidermal growth factor receptor (EGFR), a known therapeutic target. Although there are many radiomics studies for predicting disease outcomes, little is known in particular on the impact of radiomics in neuroblastoma prognosis.
The application of convolutional neural networks (CNN) is an alternative potential approach for processing medical images to evaluate patient outcomes. A CNN architecture may be trained end to end with segmented tumor scans as input and disease labels as output. Although a deep learning model like CNN depends on a large number of labeled data for training, it has been shown that transfer-learning techniques previously trained on a large unrelated diverse dataset, such as ImageNet, can reduce the need for a large number of relevant training data [12, 13]. We present a predictive model based on a pre-trained 2D CNN architecture, namely, the VGG19. We also compared the classification performance of the resulting 2D CNN model with 3D radiomic feature–based models.
Our investigation is the first study to look into the 3D radiomics–based results and compare them against 2D CNN–based results to six prognostic features in neuroblastoma with CT images. We attempt to predict histopathology and clinical outcomes. In this study, we hypothesized that computed tomography (CT)–based radiomic features have a strong relationship with tumor heterogeneity that relates to the investigated endpoint patient outcomes: mortality (presence/absence of death during the study period), the presence of image-defined risk factors (IDRF), the grade of tumor differentiation, the presence or absence of metastases, the mitosis karyorrhexis index (MKI), the amplification of the MYCN oncogene. We used a few relevant machine learning (ML) models and compared the classification performance of these models on each outcome. Nested cross-validation approach [14] to select the best set of parameters and hyper-parameters for machine-learned models. Parameters representing an ML model are used to predict at the inferencing stage. The human-selected hyper-parameters are typically pre-assigned even before training starts. Nested-cross validation allows automatic selection of hyper-parameters at the cost of extra logistics in training. Subsequently, we report appropriate models for the outcomes with respect to our small data set. Finally, we provide our perspectives on algorithms’ relative performances for each outcome.
Materials and Methods
Description of Dataset
This work was a retrospective study of medical images and records that are qualified as exempt by the appropriate Institutional Review Board (IRB). Data collected for this cohort included pretreatment CT images, and clinical and histopathology information. All clinical outcomes were collected from the electronic health records or physical medical charts. Images were anonymized before processing. Relevant patient outcomes data were abstracted from the medical records and linked to the respective images while maintaining the anonymity of the subjects. Our retrospective study was approved by the appropriate IRB. The cohort included patients enrolled at two tertiary care academic pediatric hospitals (UCSF Benioff Children's Hospital, San Francisco, and UCSF Benioff Children's Hospital, Oakland, both in California, USA) from 2000 to 2015 with pathology-proven neuroblastoma or ganglio-neuroblastoma. A total of 65 pediatric patients (age range 0–16.3 years, mean age 2.6 years, 31 males and 34 females) met inclusion criteria (Table 1). The data of 35 patients were collected at a children’s hospital (name to be included after review). The other 30 patient data were collected at another children’s hospital of the same institution but in a different location.
Table 1.
The statistics of prognostic outcomes in the neuroblastoma dataset
| Prognostic outcomes | Positive prognosis (favorable outcomes to the patient) | Negative prognosis (unfavorable outcomes to the patient) | Unknown prognosis |
|---|---|---|---|
| Presence of IDRF | No: 12 (19%) | Yes: 53 (81%) | |
| Grade of neuroblastic differentiation | Well: 55 (84%) | Poor: 7 (11%) | Unknown: 3 (5%) |
| Presence of metastases | No: 41 (63%) | Yes: 24 (37%) | |
| MKI | Low: 41 (63%) | Intermediate: 17 (26%) | Unknown: 7 (11%) |
| MYCN status | Normal: 45 (69%) | Amplified: 11 (17%) | Unknown: 9 (14%) |
| Mortality (presence/absence of death in 3 years) | No: 56 (86%) | Yes: 9 (14%) |
In this study, we investigated six prognostic patient outcomes, as mentioned before. We divided them into three primary outcomes and three secondary outcomes.
Primary Outcomes
(1) Mortality indicates death from the inception of the study in 2013 through the time of the patient’s last interaction with the health care system (for some cases up until the year 2018). (2) The second primary outcome is the presence of metastases that represents if the neuroblastoma has spread outside the primary tumor and if it can be found in other tissues on CT images. (3) Another primary outcome, the grade of neuroblastic differentiation, determines the neuroblastoma pathology differentiation, which describes the maturity of the neuroblastic cells. Poorly differentiated cells are immature and may, in combination with other disease features, indicate more aggressive tumor behavior.
Secondary Outcomes
(1) The presence of an image-defined risk factor (IDRF) is a surgical risk factor detected with imaging at the time of diagnosis [15]. The IDRF mainly evaluates if the neuroblastomas encase the surrounding vessels or organs and is an important factor in stratifying neuroblastoma. (2) The mitosis-karyorrhexis index (MKI) refers to the combined number of cells in mitosis or undergoing karyorrhexis, based on the evaluation of 5,000 tumor cells [16]. MKI results are then classified as follows: low (< 2% or < 100/5000 cells) or intermediate (> 2% and < 4% or 100 ~ 200/5000 cells) or high (> 4% or > 200/5000 cells). In this dataset, we do not have high MKI patients. The determination of the MKI involves a manual count of sufficient microscopic fields to include 5000 cells. (3) MYCN is a gene that is overexpressed in several different types of cancers, most notably in neuroblastoma. In neuroblastoma, the MYCN oncogene amplification is an established indicator of poor prognosis [17].
Segmentation and Radiomic Feature Extraction
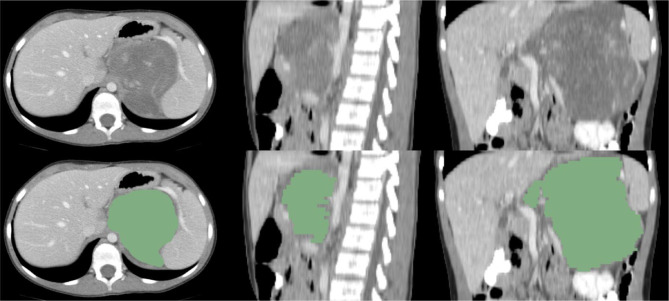
Primary tumors were hand-segmented from initial staging CT scans using the freely available open-source software package, 3D-slicer (https://www.slicer.org). A subspecialty-trained pediatric radiologist with over 8 years of experience has independently reviewed the hand-segmented primary tumors. The initial manual segmentation was performed by one of the co-authors (MP). These manual segmentations were verified by a pediatric radiologist (MZ). A neuroblastoma primary tumor segmentation example is shown in Fig. 1. The Pyradiomics library v2.2.0, as an extension of 3D-slicer, was used for extraction of radiomic features [18], which was implemented according to consensus definitions of the Imaging Biomarkers Standardization Initiative (IBSI), with a total of 105 quantitative features. No wavelet features were incorporated. We extracted all 105 3D radiomic features to characterize tumors, and these features can be categorized in the following classes: 18 first-order statistics features, 13 shape-based features, 23 Gray Level Co-occurrence Matrix (GLCM), 14 Gray Level Dependence Matrix (GLDM), 16 Gray Level Run Length Matrix (GLRLM), 16 Gray Level Size Zone Matrix (GLSZM), and 5 Neighboring Gray Tone Difference Matrix (NGTDM). The descriptions and mathematical formulas of all radiomic features are defined in the Zwanenburg review paper [19].
Fig. 1.
Original images and manual segmentations of neuroblastoma on CT image in axial, sagittal, and coronal views
In summary, the first-order statistical features capture image signal intensity and the distribution of the tumor intensity. The shape features describe the 3D geometric shape of a tumor. And the other features derived from GLCM, GLDM, GLRLM, GLSZM, and NGTDM present the spatial relationship information between neighboring voxels within the tumor region. All of these radiomic features together characterize the heterogeneity of the tumor. Radiomic features were normalized over all patients’ data to standard z-scores before building outcome classifiers, as the min–max ranges of these feature values vary. The use of z-scores, as opposed to raw scores, is a standard procedure in ML in multi-variable analysis. All 105 features are used in modeling as described in the next section.
Classifiers and Validation
We developed six ML models and a CNN model to classify the patient outcomes as listed in Table 1. The selected ML algorithms were multi-layer artificial neural network (ANN), lasso regression, elastic regression, logistic regression, random forest, and support vector machine classifier. We designed a three-layer fully connected ANN architecture containing a single hidden layer with 10 hidden units. The activation function of each neuron in the hidden layer was the sigmoid function. The momentum in the back-propagation process was set as 0.2. We hand-tuned all the hyper-parameters of the ANN, and they are as follows: learning rate = 0.00025, beta_1 = 0.9, beta_2 = 0.999, epsilon = 1e-08, and decay = 0.0. To select radiomic features, the regularization method was embedded in classifiers. For elastic regression, we used L1 and L2 regularization methods. For the Lasso regression model, we used the L1 regularization method. The hyper-parameters of other algorithms were selected by a grid search approach. Grid search is an approach that can configure optimal hyper-parameters by scanning a set of potential hyper-parameters. Nested cross-validation and grid search algorithm [14] was used to optimize the best-performed parameters. To reduce the potential overfitting, we applied a nested cross-validation approach to split the dataset into training and validation groups repeatedly. In this study, we used stratified fivefold outer cross-validation and threefold internal cross-validation to analyze the performance of our models. The grid search algorithm is a search algorithm to select the optimally performing hyper-parameters. Inner cross-validation together with grid search is used to tune the parameters in each ML model.
We used a conventional 2D CNN to predict patient outcomes directly from the CT image instead of using a 3D CNN in this study. This is primarily due to the availability of a pre-trained model in 2D. We used the fixed-size bounding box image (224 × 224) to crop the tumor out of each slice to ensure that the extracted features correspond to the same spatial information across all images. We made sure that the tumor is at the center of the bounding box. Raw images were used, and no pre-processing filter was applied. We cropped 1625 2D images from 65 neuroblastoma CT cases (25 image sample-slices per patient). We used the VGG19 model [20] to extract deep features of neuroblastoma images. The VGG19 model contains five blocks, each of which contains two or four convolutional layers and one max pooling layer. We added three fully connected layers at the end of the last convolutional layer. This model takes 2D image input to three RGB channels. In this study, the input images were duplicated across the three channels since they were in grayscale. All the hyper-parameters in the model were pre-trained by the well-known ImageNet dataset. We used VGG19’s default hyper-parameters and added a fully connected layer with two neurons at the end of the architecture. We did not perform any image augmentation, as our training set was relatively large (approximately 1600 2D-slices), and traditional geometric image transformations are not useful in this case.
For radiomic feature–based studies, the synthetic minority over-sampling technique (SMOTE) [21] was used to balance the predictive label nested within grid search during training. The area under the receiver operator characteristic curve (ROC-AUC) was selected as the model evaluation metric to quantify the predictive performance of different models. Each experiment was repeated ten times to evaluate the mean and standard deviation values of ROC-AUC. “Experiment” in this in silico context means running the cross-validation process. For the CNN model, we first calculated the mean prediction score of 25 images on each patient. We then generated the mean and standard deviation of ROC-AUC results for all fivefold testing datasets.
Results
We compared the performance of six radiomics-based ML models, over all the available 105 3D radiomic features, and one 2D CNN–based model in classifying six known patient outcomes. Model performance results are presented in Tables 2, 3, and 4.
Table 2.
Mean ROC-AUC value table of a machine learning model for three primary prognostic outcomes (bold text indicates the highest value for each outcome or row, along with the corresponding p-value indicating the significance of its difference from the next largest value in the row; standard deviation values are in parentheses next to the mean)
| Radiomics-based ANN | Lasso regression | Elastic regression | Logistic regression | Random forest | Support vector machine | ||
|---|---|---|---|---|---|---|---|
| Mortality |
0.79 (0.045) (p = 0.001) |
0.72 (0.058) |
0.72 (0.063) | 0.71 (0.041) |
0.65 (0.055) |
0.76 (0.039) | |
| Presence of metastases |
0.83 (0.034) (p = 0.011) |
0.81 (0.043) |
0.79 (0.048) |
0.68 (0.053) |
0.81 (0.028) |
0.78 (0.061) | |
| Grade of neuroblastic differentiation | 0.80 (0.047) |
0.75 (0.040) |
0.82 (0.044) (p = 0.030) |
0.79 (0.038) |
0.78 (0.051) |
0.78 (0.066) | |
Table 3.
Mean ROC-AUC value table of machine learning model versus prognostic outcomes (bold text indicates the highest value for each outcome or row; standard deviation values are in parentheses). As in Table 2, the p-value for each row’s best mean compares against the second-best performing model in the row
| Radiomics-based ANN | Lasso regression | Elastic regression | Logistic regression | Random forest | Support vector machine | ||
|---|---|---|---|---|---|---|---|
| Presence of IDRF |
0.76 (0.021) (p = 0.033) |
0.64 (0.033) |
0.75 (0.025) | 0.66 (0.045) |
0.71 (0.038) |
0.73 (0.273) | |
| MKI |
0.66 (0.031) (p = 0.011) |
0.61 (0.042) |
0.60 (0.036) | 0.64 (0.045) |
0.62 (0.047) |
0.60 (0.039) | |
| MYCN status |
0.77 (0.038) (p = 0.001) |
0.72 (0.048) |
0.73 (0.052) | 0.67 (0.031) |
0.66 (0.053) |
0.71 (0.043) | |
Table 4.
Mean ROC-AUC results of 2D CNN model. Standard deviation values are in parentheses
| Prognostic outcomes | ROC-AUC value |
|---|---|
|
Primary outcomes Mortality Presence of metastases Grade of neuroblastic differentiation Secondary outcomes Presence of IDRF MKI MYCN status |
0.76 (0.046) 0.79 (0.055) 0.77 (0.068) 0.71 (0.057) 0.63 (0.024) 0.74 (0.037) |
Statistics of Prognostic Outcomes
Table 1 shows the statistics of selected prognostic outcomes of patients in our dataset.
Radiomics-based Models
Among the ML techniques we experimented with, the multi-layer ANN model over radiomic features of 3D images outperformed the other models in predicting all the patient outcomes, except in one classification task. The results for radiomics-based analysis are shown in Tables 2 and 3.
Primary outcomes
Table 2 presents the ROC-AUC values for six ML models on tumor images. For the classification of the mortality, the overall “best performing” model ANN reached ROC-AUC values of 0.79 ± 0.045. To justify the statement, we compared this distribution against that from the support vector machine, which provides the next-best ROC-AUC value. The corresponding p-value (with independent t-test) is shown in the table. In predicting another primary patient outcome, the presence of metastases, ANN again provides the best ROC-AUC of 0.83 ± 0.034. For the third another primary patient outcome, grade of neuroblastic differentiation, the radiomics-based elastic regression approach outperformed all other approaches achieving the performance of 0.82 ± 0.044. We provided p-value for each of the “best performing” model against the second-best performing one.
Secondary outcomes
For three secondary outcomes, the best performing model, ANN, reached mean ROC-AUC values of 0.76 ± 0.021, 0.66 ± 0.031, and 0.77 ± 0.038 for the presence of image-defined risk factor (IDRF), the mitosis-karyorrhexis index (MKI), and the presence or absence of MYCN gene amplification, respectively. Table 3 shows the performance of all six ML models. The statistical significance is estimated with p-value computation, as before.
2D Image–based CNN Model
The performance of CNN on 2D slices of tumors is uniformly poorer than the best performance from other models. Table 4 presents results for all six patient outcomes. It is notable that the differences of performance are not always necessarily significant. Our observation is based on the mean values from our experiments. This ML model achieves its highest performance of 0.79 ± 0.55 for the presence of metastases.
Discussion
Discussion on Machine Learning Model
In this study, we have investigated the classification performance of multiple ML algorithms from imaging data. We analyzed 65 neuroblastoma patient data, which is a relatively large number of cases that one can obtain for this disease from a single institution. To the extent we know, there is no such study of applying ML over radiological images in neuroblastoma.
Radiomics-based Models
In Tables 2 and 3, we observe that most algorithms’ performances are relatively close to each other (within a standard deviation range), even though the radiomics-based ANN model over 3D radiomic features seems to be consistently accurate for most outcomes (within the range of 0.76 to 0.83), except for MKI (0.66). For each outcome, this method’s mean performance is the highest among all ML algorithms, except for the grade of neuroblastic differentiation, although the differences are not always substantial. However, even in the case of neuroblastic differentiation, the difference is within a standard deviation with respect to the best performing elastic regression. Similar capabilities of ANN with radiomic features have been reported within many research areas in bioinformatics, such as disease classification and identification of biomarkers [22]. We also found that the lasso regression and elastic regression models, which are linear regression models with L1 or L2 regularizations, respectively, perform better than the pure linear regression models without regularization. This is possibly because L1 or L2 regularizations are able to reduce the overfitting problem. Even though we did not reduce the number of radiomic features with any external selection method, presumably, the elastic regression’s capability to focus on important features helped it. In interpreting results reported in Tables 2 and 3, one must keep in mind the low sample size.
2D Image–based CNN Model
Compared to the 3D radiomics–based ANN model, the 2D CNN image–based model resulted in relatively poorer classification performance over our limited data set. This is likely because of the following four reasons. First, 3D radiomics had access to statistics on neuroblastoma tumors in a higher dimension than what 2D CNN had. The 3D radiomics technique is able to utilize not only voxels’ relationships within the same slice but also voxels’ relationships between neighboring slices. Second, the 2D CNN model is trained on 2D images of a rectangular box containing tissues outside the neuroblastoma tumors that may have potentially introduced noise into the training dataset, especially for our relatively small amount of data. In comparison, the 3D radiomics model is trained only on tumor segmentation without such noise. Third, due to the heterogeneity of neuroblastoma, the 2D CNN may have been trained on a slice that is not related to the outcome label for the whole tumor. This is because we used multiple slices of each 3D image over the tumor region as if each slice were independent but in reality, they had the same labels as that of the 3D image. This may result in some of the training data being potentially mislabeled. Fourth, CNN is a data-hungry technique and our sample size was not necessarily enough for it.
There are a few reasons why we (and often other researchers on similar problems) chose to use 2D CNN rather than 3D CNN. (1) As mentioned before, usage of pre-trained VGG19 required 2D images as input. Pre-trained models reduce the need for an even larger training data set and provide better accuracy than otherwise. We hope that the availability of pre-trained 3D CNN models in the future will alleviate this restriction in medical imaging research but presently, we had to accept this limitation. (2) Training with 3D images was computationally prohibitive, even using an NVIDIA V-100 GPU. (3) Working with the 2D slices of the 3D images, rather than the 3D images themselves, increased our training and testing data size by many folds (× 25) than what we would have had with original 3D images. As noted earlier, our training set is relatively small for the purpose of using CNN. This approach of using 2D slices where the number of available 3D images is not enough is often discussed in medical imaging research. Small sample size did not pose a significant challenge for other ML models that used 3D radiomics information as in the CNN model since the curse of dimensionality is much less with the former. The dimension or number of radiomic features is slightly more than one hundred. In contrast, the dimension for a 2D image is the number of pixels that runs close to thousands, even after using only the masked region of interest.
Discussion on Patient Outcome Prediction
Primary Outcomes
Among all outcomes, the radiomics-based ANN model was able to classify the presence of metastases with the highest ROC-AUC (0.83 ± 0.034). In general, we found that CT radiomic features have good predictability of the presence of metastases and neuroblastic differentiation (0.82 ± 0.044). A similar high correlation between the presence of metastases and CT radiomic features can also be found in many other studies [23–25]. The success of predicting the mortality is quite surprising, where the highest ROC-AUC value is achieved from the ANN: 0.79 ± 0.045. If similar performance is observed with a larger sample size in the future, the model parameters themselves may be investigated to provide deeper insight on how mortality rate is being correlated with radiomic features.
Secondary Outcomes
The presence of image-defined risk factors (IDRFs) in neuroblastic tumors depends on the severity of primary neuroblastoma encasing vessels or invading neighboring organs. The presence of IDRFs has a strong correlation with patients’ prognoses. It also helps to predict surgical outcomes, provides risk stratification, and guides international neuroblastoma risk group’s (INRG’s) staging system [26]. However, primary tumor heterogeneity is not directly related to IDRFs. Thus, our models perform moderately (max ROC-AUC 0.76 from radiomics-based ANN) in predicting the presence of IDRFs. Our study also suggested that radiomic features may have lesser information content regarding the presence of MKI than for predicting other outcomes. Similar reasons may account for the intermediate performance for predicting mortality and amplified MYCN outcomes. MYCN outcome indicates the amplification of the MYCN gene. According to Gillies et al. [27], radiomic features can provide important information regarding the sample genomics but are not significantly related to gene expression, which has been proved to be helpful in the cases of neuroblastoma [28].
We found that MKI prediction is low across all ML algorithms (0.60–0.66), and actually, the ANN prediction value is the highest (0.66) in the respective row in Table 2. Low prediction accuracies across all algorithms indicate that the observer-dependent MKI is not well-predicted by ML models based on CT images. We believe its low prediction capability (0.66) for MKI is because CT images do not have enough resolution to predict the mitosis and karyorrhexis that this index measures. MKI indicates that a pathologist counted 5000 or more cells on the neuroblastoma patient’s histology under the microscope, which is time-consuming and observer dependent. This makes MKI labels somewhat subjective. Literature has suggested an alternative approach to reduce observer dependence in measuring the MKI [20]. Similar to Atikankul’s finding [20], our result also indicates that the observer-dependent MKI may not be accurate and is not as reliable as other outcomes.
Conclusion
Our study provides a proof of concept that ML over previously extracted radiomic features may be a better alternative than a 2D CNN that is trained over slices of radiological images to predict patient outcomes. However, we could not perform a comparison against 3D CNN for computational limitations, which may be the case in many practical scenarios. The approach of training over radiomic features is computationally much easier to perform. Given the concern within the community on the variability of radiomic features across different studies, our result from data spanning two different hospitals is somewhat interesting, even though both hospitals are under the same institution using the same vendor’s machines with the same protocol for imaging. It is to be noted that the radiomic features are normalized (z-score) before being used by ML algorithms. However, we could not study the cohorts independently to validate the independence of radiomic features for small sample sizes in each of the cohorts. Hopefully, our work will inspire a broader multi-institutional study to validate this. The small sample size and the possibility of feature variation remain limitations in our study.
It is also worth investigating the role of individual radiomic features in “explaining” the outcomes, whereas a CNN applied directly on images has limited explanation capability. As future directions of this work, similar studies with multiple ML models (over radiomic features or directly on images) may be conducted over other medical imaging areas (disease models and imaging modalities) to converge on the appropriate models for clinical use. Given the limited size of data, we did not investigate relative importance of features and model parameters. We intend to do so in the future to gain possible key insights [29]. Finally, given the small sample size, the generalizability of our conclusions remains a limitation in our study.
In this paper, we decided to present seven ML models’ performances that produced the best results out of a few different techniques we tried. For any clinical deployment, it may be necessary to use multiple ML models’ results and use a weighted voting approach to provide a final prediction for each outcome along with a confidence measure.
Potent future work is to automate the tumor segmentation process and then perform the radiomics-based analysis or apply CNN. This automation will make our work feasible to be verified on a large-scale dataset where manual segmentation of each tumor is not scalable over a study with a large sample size. Automated segmentation will also make the clinical deployment of ML models easier.
Acknowledgements
This work was supported in part by the National Cancer Institute Grant R01CA154561 and the National Institute of Biomedical Imaging & Bioengineering Grant R15EB030807. Anonymous reviewers’ comments have significantly improved the article.
Declarations
Ethics Approval
This study was a retrospective study of medical records and medical images and qualified as exempt by the appropriate Institutional Review Board (IRB) at the Florida Institute of Technology.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Debasis Mitra, Email: dmitra@cs.fit.edu.
Youngho Seo, Email: youngho.seo@ucsf.edu.
References
- 1.Maris John M, Matthay Katherine K. Molecular biology of neuroblastoma. Journal of Clinical Oncology. 1999;17(7):2264–2264. doi: 10.1200/JCO.1999.17.7.2264. [DOI] [PubMed] [Google Scholar]
- 2.Maris JM, Hogarty MD, Bagatell R. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 3.Caron HN. Are thoracic neuroblastomas really different? Pediatric Blood & Cancer. 2010;7(54):867–867. doi: 10.1002/pbc.22470. [DOI] [PubMed] [Google Scholar]
- 4.Goodman MT, Gurney JG, Smith MA, Olshan AF. “Sympathetic nervous system tumors. Cancer Incidence and Survival among Children and Adolescents.” United States SEER Program, 65–72 (1995).
- 5.Jereb B, Bretsky SS, Vogel R, Helson L. Age and prognosis in neuroblastoma. Review of 112 patients younger than 2 years. The American Journal of Pediatric Hematology/Oncology. 1984;6(3):233–243. doi: 10.1097/00043426-198423000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Kulkarni AV, Bilbao JM, Cusimano MD, Muller PJ. Malignant transformation of ganglioneuroma into spinal neuroblastoma in an adult: case report. Journal of Neurosurgery. 1998;88(2):324–327. doi: 10.3171/jns.1998.88.2.0324. [DOI] [PubMed] [Google Scholar]
- 7.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, Van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. European Journal of Cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teshiba R, Kawano S, Wang LL, He L, Naranjo A, London WB, Seeger RC, Gastier-Foster JM, Look AT, Hogarty MD, Cohn SL. Age-dependent prognostic effect by Mitosis-Karyorrhexis Index in neuroblastoma: a report from the Children’s Oncology Group. Pediatric and Developmental Pathology. 2014;17(6):441–449. doi: 10.2350/14-06-1505-OA.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson A, O’Connor JP, Parker GJ, Jayson GC. Imaging tumor vascular heterogeneity and angiogenesis using dynamic contrast-enhanced magnetic resonance imaging. Clinical Cancer Research. 2007;13(12):3449–3459. doi: 10.1158/1078-0432.CCR-07-0238. [DOI] [PubMed] [Google Scholar]
- 10.Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, Aldape K, Cha S, Kuo MD. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proceedings of the National Academy of Sciences. 2008;105(13):5213–5218. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang SY, Franc BL, Harnish RJ, Liu G, Mitra D, Copeland TP, Arasu VA, Kornak J, Jones EF, Behr SC, Hylton NM. Exploration of PET and MRI radiomic features for decoding breast cancer phenotypes and prognosis. NPJ Breast Cancer. 2018;4(1):24. doi: 10.1038/s41523-018-0078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donahue J, Jia Y, Vinyals O, Hoffman J, Zhang N, Tzeng E, Darrell T. “Decaf: a deep convolutional activation feature for generic visual recognition.” International Conference on Machine Learning 647–655, (2014).
- 13.Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Transactions on Medical Imaging. 2016;35(5):1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cawley GC, Talbot NL. “On over-fitting in model selection and subsequent selection bias in performance evaluation.” Journal of Machine Learning Research 2079–107, (2010).
- 15.Atikankul T, Atikankul Y, Santisukwongchote S, Marrano P, Shuangshoti S, Thorner PS. MIB-1 index as a surrogate for mitosis-karyorrhexis index in neuroblastoma. The American Journal of Surgical Pathology. 2015;39(8):1054–1060. doi: 10.1097/PAS.0000000000000478. [DOI] [PubMed] [Google Scholar]
- 16.Gestblom C, Hoehner JC, Påhlman S. Proliferation and apoptosis in neuroblastoma: subdividing the mitosis-karyorrhexis index. European Journal of Cancer. 1995;31(4):458–463. doi: 10.1016/0959-8049(95)00006-5. [DOI] [PubMed] [Google Scholar]
- 17.Yoshimoto M, Caminada De Toledo SR, Monteiro Caran EM, et al. “MYCN gene amplification. Identification of cell populations containing double minutes and homogeneously staining regions in neuroblastoma tumors.” Am J Pathol. 155(5):1439‐1443, (1999). 10.1016/S0002-9440(10)65457-0 [DOI] [PMC free article] [PubMed]
- 18.Van Griethuysen JJ, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RG, Fillion-Robin JC, Pieper S, Aerts HJ. Computational radiomics system to decode the radiographic phenotype. Cancer Research. 2017;77(21):104–107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zwanenburg A, Leger S, Vallières M, Löck S, et al. Image biomarker standardisation initiative. Radiology. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonyan K, Zisserman A. “Very deep convolutional networks for large-scale image recognition.” arXiv preprint arXiv:1409.1556. 2014 Sep 4.
- 21.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. Journal of Artificial Intelligence Research. 2002;16:321–357. doi: 10.1613/jair.953. [DOI] [Google Scholar]
- 22.Lancashire LJ, Lemetre C, Ball GR. An introduction to artificial neural networks in bioinformatics—application to complex microarray and mass spectrometry datasets in cancer studies. Briefings in Bioinformatics. 2009;10(3):315–329. doi: 10.1093/bib/bbp012. [DOI] [PubMed] [Google Scholar]
- 23.Coroller TP, Grossmann P, Hou Y, Velazquez ER, Leijenaar RT, Hermann G, Lambin P, Haibe-Kains B, Mak RH, Aerts HJ. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiotherapy and Oncology. 2015;114(3):345–350. doi: 10.1016/j.radonc.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang YQ, Liang CH, He L, Tian J, Liang CS, Chen X, Ma ZL, Liu ZY. “Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer.” Jnl. of Clinical Oncology 34(18), (2016). [DOI] [PubMed]
- 25.Chen AM, Trout AT, Towbin AJ. A review of neuroblastoma image-defined risk factors on magnetic resonance imaging. Pediatric Radiology. 2018;48(9):1337–1347. doi: 10.1007/s00247-018-4117-9. [DOI] [PubMed] [Google Scholar]
- 26.Brisse HJ, McCarville MB, Granata C, Krug KB, Wootton-Gorges SL, Kanegawa K, Giammarile F, Schmidt M, Shulkin BL, Matthay KK, Lewington VJ. Guidelines for imaging and staging of neuroblastic tumors: consensus report from the International Neuroblastoma Risk Group Project. Radiology. 2011;261(1):243–257. doi: 10.1148/radiol.11101352. [DOI] [PubMed] [Google Scholar]
- 27.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2015;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brisse HJ, et al. Radiogenomics of neuroblastomas: relationships between imaging phenotypes, tumor genomic profile and survival. PLOS One. 2017 doi: 10.1371/journal.pone.0185190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu G, Mitra D, Jones EF, et al. Mask-guided convolutional neural network for breast tumor prognostic outcome prediction on 3D DCE-MR images. J Digit Imaging. 2021 doi: 10.1007/s10278-021-00449-y. [DOI] [PMC free article] [PubMed] [Google Scholar]