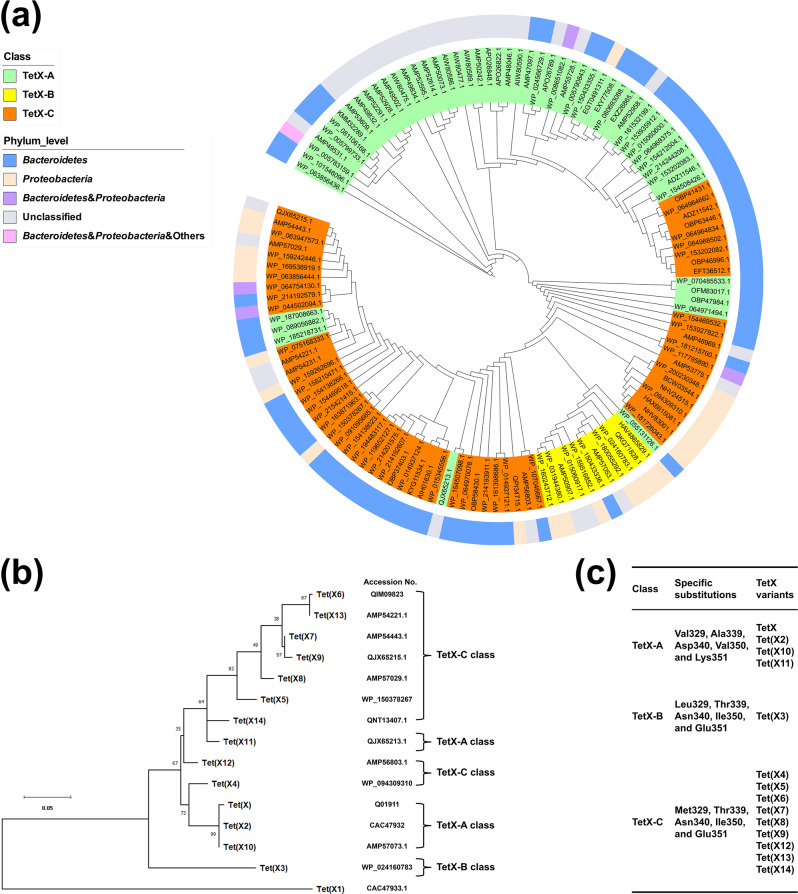
Fig. 3. Phylogenetic analysis of TetX like protein.
a TetX-related proteins are divided into three classes: TetX-A (green), TetX-B (yellow), and TetX-C (orange). Phylogeny is inferred by using the maximum-likelihood method and Flu +G + I model. A discrete Gamma distribution approach was used to depict the difference in evolutionary rate among the sites [4 categories (Gamma shape parameter = 0.609)]. This analysis involved 128 amino acid sequences. Different species hits the protein sequences organized in the phylum level. Bacteroidetes, Proteobacteria, Firmicutes, Spirochetes, Bacteroidetes plus Proteobacteria, and unclassified organisms are shown in blue, pink, light purple, and gray, respectively. Evolutionary analyses were conducted in online PlyML 3.0. The tree was visualized using iTOL (ITEREACTIVE TREE OF LIFE). b Phylogenetic analysis of the amino acid sequences of the reported TetX variants. The maximum-likelihood tree was inferred using MEGA X35. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. c Reported TetX variants are distributed in TetX-A class, TetX-B class, and TetX-C class with specific substitutions.