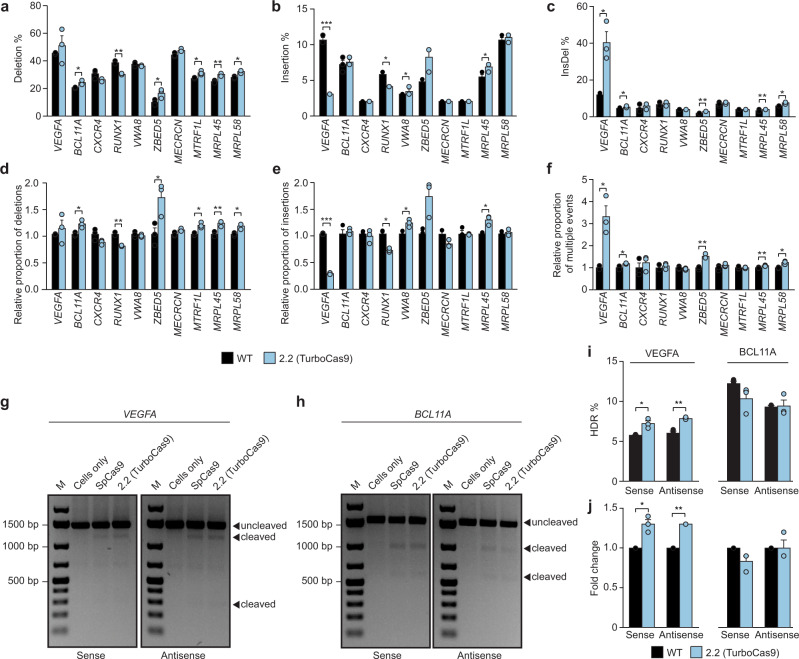
Fig. 4. An engineered Cas9 variant is highly effective at generating deletions and multiply edited complex events.
a–c Occurrence of editing events classified as a deletions b insertions c multiply edited complex mutations. d–f Relative occurrence of d deletions e insertions f multiply edited complex events. g PstI digestion of VEGFA HDR samples. M denotes the molecular weight marker. h PstI digestion of BCL11A HDR samples. i Occurrence of HDR in percentage. j Fold change of HDR. Error bars: s.e.m. for n = 3 biologically independent replicates. A standard Student’s t-test with a two-tailed distribution and unequal variance assumed between samples was used to calculate the significance. p-values: *p < 0.05, **p < 0.01, ***p < 0.001. Specific p-values for a for TurboCas9 vs wild-type Cas9: BCL11A p = 0.02, RUNX1 p = 0.005, ZBED5 p = 0.04, MTRF1L p = 0.02, MRPL45 p = 0.008 and MRPL58 p = 0.02; for b: VEGFA p = 7.0e-07, RUNX1 p = 0.02, VWA8 p = 0.02 and MRPL45 p = 0.02; for c: VEGFA p = 0.04, BCL11A p = 0.04, ZBED5 p = 0.003, MRPL45 p = 0.009 and MRPL58 p = 0.02 (FDR-adjusted Student’s t-test). Specific p-values for d for TurboCas9 vs wild-type Cas9: BCL11A p = 0.02, RUNX1 p = 0.005, ZBED5 p = 0.04, MTRF1L p = 0.02, MRPL45 p = 0.008 and MRPL58 p = 0.02; for e: VEGFA p = 7.0e-07, RUNX1 p = 0.02, VWA8 p = 0.02 and MRPL45 p = 0.02; for f: VEGFA p = 0.04, BCL11A p = 0.04, ZBED5 p = 0.003, MRPL45 p = 0.009 and MRPL58 p = 0.02 (FDR-adjusted Student’s t-test). Specific p-values for i: sense oligonucleotide p = 0.05 and antisense oligonucleotide p = 0.01. Specific p-values for j: sense oligonucleotide p = 0.05 and antisense oligonucleotide p = 0.01 (FDR-adjusted Student’s t-test). Source data are provided as a Source Data file.