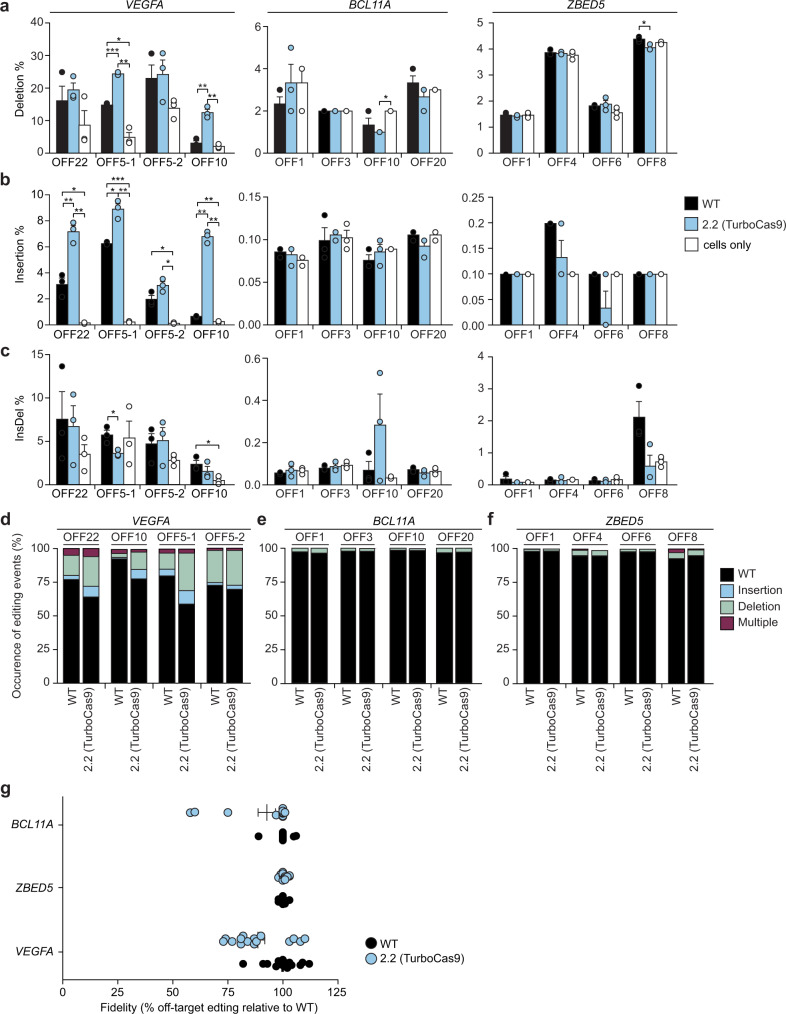
Fig. 5. Off-target DNA editing in mammalian cells.
a–c Percentage of reads for VEGFA, BCL11A and ZBED5 that have a deletions b insertions and c combined insertions and deletions (Insdel). d–f The occurrence of different editing events varies between Cas9 variants and off-target sites for d VEGFA e BCL11A and f ZBED5. g Overall fidelity of engineered Cas9 relative to wild-type Cas9 for BCL11A, ZBED5 and VEGFA, calculated as off-target error rate as a proportion of wild-type error rate. Error bars: s.e.m. for n = 3 biologically independent replicates. An FDR-adjusted Student’s t-test with a two-tailed distribution and unequal variance assumed between samples was used to calculate the significance. p-values: *p < 0.05, **p < 0.01, ***p < 0.001. Specific p-values for a for TurboCas9 vs wild-type Cas9: VEGFA OFF5-1 p = 0.00002, OFF10 p = 0.002, for ZBED5 OFF8 p = 0.02; cells only vs wild-type Cas9: VEGFA OFF 5-1 p = 0.02, BCL11A OFF8 p = 0.04; cells only vs TurboCas9: VEGFA OFF 5-1 p = 0.005, OFF10 p = 0.004, BCL11A OFF10 p = 0.02 (FDR-adjusted Student’s t-test). Specific p-values for b for TurboCas9 vs wild-type Cas9: VEGFA OFF22 p = 0.004, OFF5-1 p = 0.02, OFF10 p = 0.001; cells only vs wild-type Cas9: VEGFA OFF 5-1 p = 0.00002, OFF22 p = 0.03, OFF5-2 p = 0.03, OFF10 p = 0.002; cells only vs TurboCas9: VEGFA OFF 5-1 p = 0.002, OFF22 p = 0.004, OFF10 p = 0.001, OFF5-2 p = 0.01 (FDR-adjusted Student’s t-test). Specific p-values for c cells only vs wild-type Cas9 for VEGFA OFF10 p = 0.03; TurboCas9 vs wild-type Cas9: OFF22 p = 0.05 (FDR-adjusted Student’s t-test).