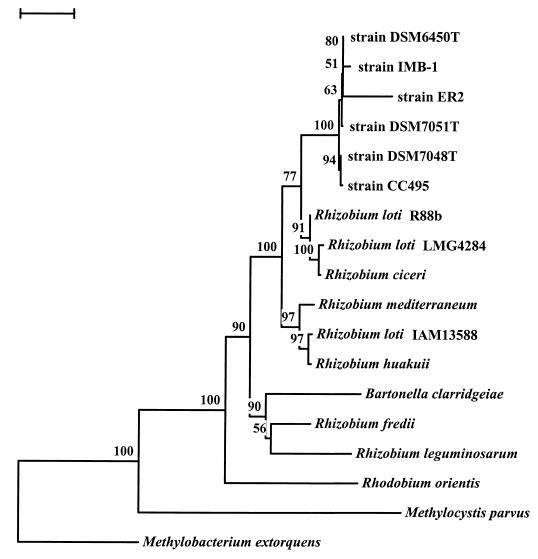
FIG. 2.
Phylogenetic analysis of the 16S rRNA gene from strain CC495. The tree was obtained by the neighbor-joining approach using the TREECON program. Similar phylogenesis was obtained when parsimony analysis of the same data was conducted. Bootstrap values (in percentages) are given at the nodes. Bar, 0.02 base substitutions per site. The 16S rRNA gene sequence from Methylocystis parvus OBBp was used as the outgroup. The GenBank accession numbers of the organisms are as follows: strain CC495, AF107722; strain DSM 7048T, AF011759; strain DSM 7051T, AF011760; strain DSM 6450T, AF011762; strain ER2, L20802; strain IMB-1, AF034798; Rhizobium loti R88b, U50165; R. loti LMG4284, X67230; Rhizobium ciceri UPM-Ca7, U07934; Rhizobium mediterraneum UPM-Ca36, L38825; R. loti IAM13588, D12791; Rhizobium huakuii IAM14158, D12797; Bartonella clarridgeiae CIP104772, X97822; Rhizobium fredii LMG6217, X67321; Rhizobium leguminosarum USDA2370, U29386; Rhodobium orientis MB312, D30792; M. extorquens, M29027; M. parvus OBBp, M29026.