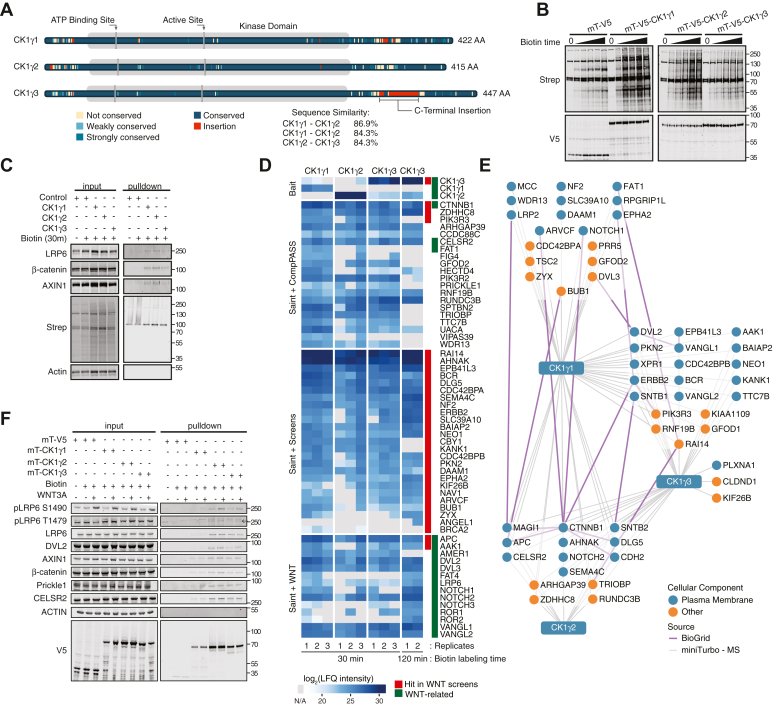
Figure 1.
CK1γ proximity-based interaction networks identify WNT components.A, schematic of CK1γ family highlighting conserved amino acids and sequence similarity between family members. B, W. blot analysis of HEK293T cells stably expressing mini-Turbo CK1γ or V5 control incubated with 50 mM of biotin over a time course (untreated, 15 min, 30 min, 45 min, 1 h, and 2 h). C, W. blot analysis of streptavidin affinity pulldown from stable HEK293T cells expressing mini-Turbo CK1γ or V5, treated as indicated. D, heat map demonstrating changes in log2(LFQ intensity) of protein proximity partners for the CK1γ subfamily. Red track indicates hits in previously published WNT screens (Tables S1 and S2). Green track indicates well-established proteins involved in WNT/β-catenin-dependent and WNT/β-catenin-independent signaling. E, streptavidin affinity purification mass spectrometry (APMS) proximity networks for CK1γ1, CK1γ2, and CK1γ. HEK293T cells stably expressing mT-V5-CK1γ were treated with 50 mM biotin for 30 min before streptavidin APMS. Blue circle indicates localization to the plasma membrane. Gray lines represent bait–prey interactions. Purple lines indicate prey–prey interactions collected from the BioGRID database. F, W. blot validation of mT-CK1γ protein–protein interaction networks treated with biotin and WNT3A CM for 30 min. CK1γ, casein kinase 1γ; CM, conditioned media; HEK293T, human embryonic kidney 293T cell line; LFQ, label-free quantitation; W. blot, Western blot.