Summary
Background
Estimating tumor purity is especially important in the age of precision medicine. Purity estimates have been shown to be critical for correction of tumor sequencing results, and higher purity samples allow for more accurate interpretations from next-generation sequencing results. Molecular-based purity estimates using computational approaches require sequencing of tumors, which is both time-consuming and expensive.
Methods
Here we propose an approach, weakly-supervised purity (wsPurity), which can accurately quantify tumor purity within a digitally captured hematoxylin and eosin (H&E) stained histological slide, using several types of cancer from The Cancer Genome Atlas (TCGA) as a proof-of-concept.
Findings
Our model predicts cancer type with high accuracy on unseen cancer slides from TCGA and shows promising generalizability to unseen data from an external cohort (F1-score of 0.83 for prostate adenocarcinoma). In addition we compare performance of our model on tumor purity prediction with a comparable fully-supervised approach on our TCGA held-out cohort and show our model has improved performance, as well as generalizability to unseen frozen slides (0.1543 MAE on an independent test cohort). In addition to tumor purity prediction, our approach identified high resolution tumor regions within a slide, and can also be used to stratify tumors into high and low tumor purity, using different cancer-dependent thresholds.
Interpretation
Overall, we demonstrate our deep learning model's different capabilities to analyze tumor H&E sections. We show our model is generalizable to unseen H&E stained slides from data from TCGA as well as data processed at Weill Cornell Medicine.
Funding
Starr Cancer Consortium Grant (SCC I15-0027) to Iman Hajirasouliha.
Keywords: Deep Learning, Computational pathology, Tumor purity estimation, Precision medicine
Research in context.
Evidence before study
Estimating the amount of tumor content in a slide has previously been performed by trained pathologists. Increasingly, computational tools have been developed to measure tumor purity, but these require sequencing, which is time consuming and expensive. Previous literature has shown that deep learning models can identify key characteristics of histological tissue slides, called H&E slides, used by pathologists to estimate tumor purity.
Added value of this study
We present a deep learning approach trained on data that is readily available at all medical institutions, these H&E stained slides, to predict tumor purity. In addition, we remove the need for sequencing of tumors to estimate this purity score.
Implications of all the available evidence
This approach allows for proper correction of sequencing data in the context of precision medicine, as well as the potential to improve understanding of clinical outcomes.
Alt-text: Unlabelled box
Introduction
In recent years, there has been an increase in tumor DNA sequencing from cancer patients, to identify tumor driving somatic mutations. However, samples that are used for sequencing consist of not only cancer cells, but also include normal immune cells, stromal cells and blood vessels, whose DNA also gets sequenced. The purity of tumor samples impacts sequencing results, making it harder to detect mutations, especially subclonal events due to dilution of tumor DNA by normal DNA. Determining tumor purity is therefore critical before sequencing. Moreover, purity in some cases can provide prognostic information. In glioma and colon cancer, low tumor purity is associated with worse survival outcomes.1,2 Therefore, tumor purity estimates can help correct sequencing outputs and help predict disease outcomes.
Traditionally, tumor purity has been estimated by pathologists based on review of hematoxylin and eosin (H&E) stained slides. Several methods, including ABSOLUTE,3 ESTIMATE,4 as well as a consensus purity estimate (CPE),5 have aimed to determine tumor purity through computational analysis of sequencing results. Molecularly derived purity scores, especially those relying on DNA, are attractive because they are thought to be highly accurate and do not need any manual review or inspection. However, sequencing and associated analysis are expensive processes, especially if tumor sample quality is not sufficient for proper analysis. On the other hand, pathologist-derived purity scores may have limited accuracy because they may have substantial inter-pathologist variability; indeed previous literature has shown that pathology-estimated tumor purity often fail to correlate well with sequencing-derived purity.6 Consensus-based approached have been one solution proposed for this problem, but they require either multiple pathologists or multiple methodologies for molecular approaches, increasing monetary and time costs.6 Thus, other approaches to quantify tumor purity are necessary.
H&E-stained slides are cheap and quick to produce from tumor samples and have been the gold standard for cancer diagnosis by trained pathologists. The utilization of artificial intelligence (AI) methods, in particular deep learning, for analyzing histopathology images has significantly increased over the past few years. Notable deep learning methods were developed to identify the presence of tumor in a tissue, segment regions of interest, and classify molecularly-derived subtypes.7, 8, 9, 10, 11, 12, 13, 14, 15, 16 There have been two main approaches to handling these types of deep learning analyses, fully-supervised and weakly-supervised. A fully-supervised approach requires each image to have a label associated with it to train a model. In fully supervised approaches, such as the data that was presented for the CAMELYON16 and CAMELYON17 datasets,17 expert pathologists will review all slides used for training and testing an algorithm and manually annotate the slides to represent classes or segmented regions of interest. Due to computational constraints, entire gigapixel sized images, with several thousand pixels in height and width, cannot be directly used as input into standard deep learning techniques, although sophisticated approaches have been proposed to make these large images compatible.18 Instead, the consensus approach to date has been to split the slides into small patches or tiles (hundreds of pixels for width and height), that can be efficiently fed into these models. In this scenario, annotated regions are necessary as there may be small sections of the tissue, at the size of a single patch, that have no tumor tissue within it. It is possible to train a model using slide level labels for patches, but this will result in noisy labels, as some non-tumor patches within a given tumor slide will be given the wrong label. The problem with a fully-supervised approach, in which manual annotations of regions is needed, is that it requires a significant amount of time from domain experts to label enough slides to be used to properly train deep learning models. Therefore, recent works have focused on using weakly-supervised approaches.
In a weakly-supervised approach, instead of manually annotating the entire slide, region by region or patch by patch, the slide is given a single overall label, and deep learning models use this information to identify regions of interest or use this to classify the disease state of the slide as a whole. This allows a much quicker annotation process, as the entire slide can be given a label and the heterogeneity within the slide can be accounted for. The main premise of most weakly-supervised approaches for histological analysis requires pooling the features from image patches, under the multiple instance learning (MIL) framework.19 In this scenario, a “bag” represents the entity in which the classification occurs, and “instances” represent heterogeneous elements that all relate to the bag entity. In the case of the standard assumption for MIL, if at least one instance within the bag is positive, the whole bag can be considered positive.19 This concept was applied by Campanella et al.,8 where the authors used > 15,000 cancer patients with several years’ worth of histological slides from the Memorial Sloan Kettering Cancer Center (MSKCC) slide repository in addition to external patient slides to predict the presence of cancer within a slide. The pooling operation used in this approach was max pooling. In this case, a slide is positive if at least one patch is predicted as positive (positive would be the presence of tumor). The authors provided a proof-of-concept study that clinical-grade performance (described as an AUC of greater than 0.98 for all cancer types for cancer detection) was achieved using this approach, and that specific malignant regions within slides were identified.8 As mentioned by Lu et al, this requires a lot of data, since very few patches from each slide are used for the classification; in fact, only one patch per slide is used during backpropagation for the max pooling operator.15 Another pooling operation that can be used is average pooling under the collective assumption of the MIL algorithm. In this regard, all of the instances within a bag, have an equal influence on the bag class.19 A seminal work was introduced in 2018, called Attention-MIL, which introduced a new pooling approach and is a modification of this collective assumption.20 The principle of this approach is that a neural network is used within the model to learn a good aggregating function. In this sense, it is learning a weighted sum of all the instances within a bag based on the feature representation of that instance, to make the prediction of interest. The different weights can then be interpreted the level of influence a particular region has on model prediction. This approach has been used to find slides that contain cancer and identify cancer subtypes,15 as well as to automatically identify the hormonal status in breast cancer from H&E slides,21 stage of cancer,22 and tissue origin of metastatic lesions.23
In this study, we propose a weakly-supervised approach to calculating tumor purity from whole slide sections utilizing an attention-based, multi-task, multiple instance deep learning model, we call wsPurity, for whole-slide purity detection. Most of the previous work done using weakly-supervised methods have provided labels to perform binary or multi-class classification, indicating 1 for the correct class label and 0 for the rest. Previous work has incorporated tumor purity in the prediction task,24 however this was performed in a fully-supervised manner. In this work, we propose that incorporating tumor purity into the classification task will allow for the proper identification of tumor regions within the histological slide, and improved performance over this fully-supervised approach.
We use an Attention-MIL setup to learn a weight feature for the distribution of patches within a slide and a feature representation that can accurately predict tissue type as well as tumor purity level. We adapt a similar deep learning pipeline to the multi-task multiple instance learning approach used in Lu et al.,23 but the fully-connected layers, specific tasks, and loss functions applied differs from their approach. Framing the purity score prediction as an ordinal classification problem, wsPurity predicts tumor purity within 9 different bins ranging from 0 to 1, where 0 is no tumor and 1 is complete tumor. We use a pathologist derived consensus purity score developed from previous literature as the ground-truth to train our model based on TCGA database slides.24 There are several unique benefits for our model, which include (1) We can accurately identify tumor purity in a tissue slide and compare our results to previously developed models, (2) We can classify tumors into low and high purity at several different thresholds that can be cancer type specific, and (3) we can identify potential tumor regions that can be isolated and used to enrich the tumor sample for improved sequencing.
Methods
Data Preprocessing
Data was acquired from The Cancer Genome Atlas (TCGA) database, and whole slide images (WSIs) were exported for downstream analysis as svs file format. 5390 slides were taken from six different tumor types and a total of 3240 total patients. These tumor types were chosen to span the range of tumor purity present in the TCGA database based on previous literature.5 We analyzed adrenal adenocarcinoma (ACC), lung squamous cell carcinoma and lung adenocarcinoma (LUSC & LUAD respectively), breast invasive carcinoma (BRCA), head and neck squamous cell carcinoma (HNSC), prostate adenocarcinoma (PRAD), and low and high grade serous carcinomas (ovarian serous cystadenocarcinoma labeled in TCGA or OV). A small number of urothelial bladder carcinoma (BLCA) normal solid tissue slides (22 slides) were included in the training set, however, we did not include any BLCA patients in the validation and test set, and there were no tumor tissue from BLCA patients in the training, validation, and test sets to be evaluated, so we do not report any metrics on this cancer type. Each tumor type was preprocessed separately. Slides were tiled into 512×512 patches at 20x resolution (adjusted field of view and downsampling if 40x magnification was used). A color threshold, empirically set for red, green, and blue, was set for each patch to identify the amount of tissue present, and a threshold was set at 40% tissue presence to remove background regions. Color thresholding was performed by converting the image into HSV color space and performing a threshold using the opencv-python package. A Haar wavelet transform was used to filter out tissue that was out of focus due to issues related to the scanner.32,33 An empirically derived threshold was set for the blur detection to minimize loss of tissue while still removing severely out of focus images (41,632 out of 3,730,063 total patches or ∼1.1%). The slides were split for each tumor such that there was approximately 70% | 15% | 15% for train | validation | test sets for each tissue (Supplemental Table 1). A small fraction of tissues in the training set had less than N=120 number of tiles in the slide and were removed from the analysis during training. Slides were split so that each tissue sample was split into one category. Therefore, for cancer samples (-01A sample code labeled through TCGA), all slides from the same patient were grouped into the same split, and for each normal sample (-11A sample code), the slides from a single patient were grouped into the same split. Data distributions as well as predictions for the validation and test sets can be viewed in the supplemental materials (Supplemental Tables 2–4).
In addition, we used an WCM independent cohort (outside of TCGA) to validate our model perform. We received 78 de-identified patient frozen section H&E slides from 48 different patients, from three different cancer types, BRCA and PRAD, with a pathologist-derived purity estimate to compare with our proposed method.
Model
We used Pytorch for this study (version 1.1)34 and utilized a variant of Resnet34 that is called Resnet34-IBN,25 which had initially been trained on ImageNet,36 and adds InstanceNorm35 (preprint) layers within the model (Figure 1c). In histology images there are noticeable color variations due to differences in tissue preprocessing and stain protocols. InstanceNorm has been used to filter complex color variations (i.e. color shifts or brightness changes), and was utilized for that reason, instead of changing the input of the image through methods such as stain normalization done in other studies.7,25 To improve model performance and minimize overfitting, we added several different regularization methods. We have added weight decay (L2 Norm) regularization (1×10−4), data augmentation (imgaug package) including rotations/flips, coarse dropout, Gaussian noise, hue/saturation/contrast adjustment, and intensity scaling. We use a learning rate of 0.005, the stochastic gradient descent (SGD) optimizer, and a bag size of 120 patches, with a batch size of 2 for the MIL set-up. We tested a bag size:batch size ratio of 60:4, 120:2, and 300:1, and the 120:2 gave the best results and therefore were used for all analyses (data not shown). Multi-task learning was performed to analyze both the tissue type present in the slide as well as tumor purity. For tissue type prediction cross-entropy loss was used with inverse frequency of each class used to weight each component in the loss function. For tumor purity the loss used was derived from a previous study, and is used to be able to introduce an ordering to the classification task.37 Using this ranked loss function, a probability distribution (, where is the threshold used to separate class from class (where in our case is 8) is generated for each threshold. The additional class, in addition to the 8 that fall above each of the i thresholds, is formed for samples that are below all thresholds (i.e. less than 0.09 or less than 9% pure). We binned tumor purity using the following thresholds [0.09, 0.29, 0.39, 0.49, 0.59, 0.69, 0.79,0.89], such that a tumor purity of 0 represents normal tissue (denoted as -11A in the TCGA database) and 1 represents pure tumor. We removed the 0.19 threshold due low representation (14 WSI examples out of 5,860 in the entire dataset or < 0.1% of the total number of slides) within this class. Through this approach, we can set 8 different thresholds when classifying tumors into high vs. low purity, where different thresholds can be used based on biological significance.
Figure 1.
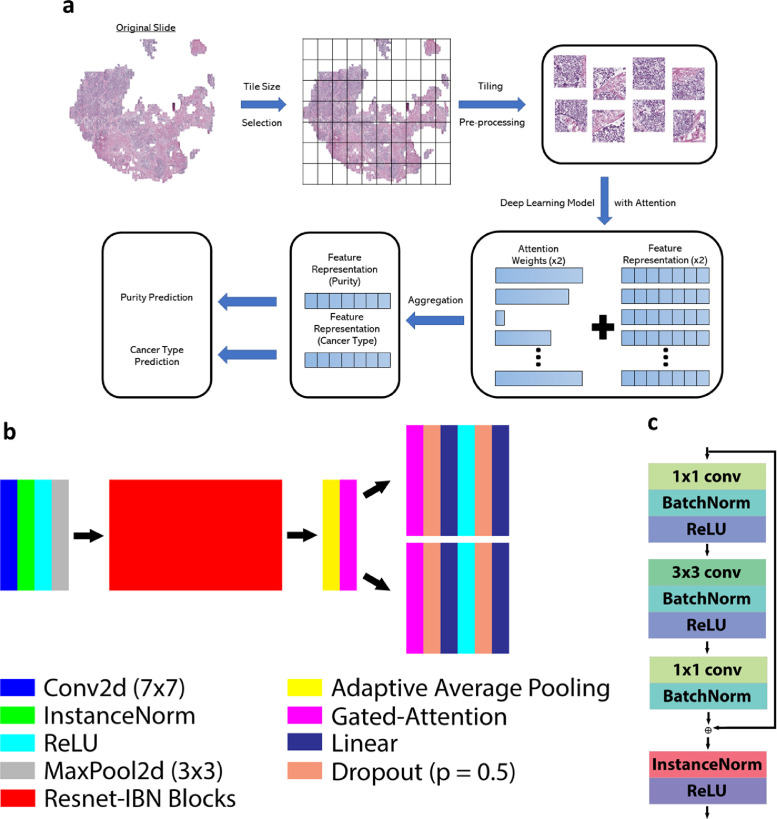
(a) Workflow of wsPurity. To get a slide output, the original svs file is tiled, passed through a deep learning model with an attention mechanism to combine information from all tiles to perform two tasks, cancer type prediction and tumor purity prediction. (b) Schematic of the multi-attention multi-task MIL approach. The model uses the structure of Resnet-34-IBN, which is a modified Resnet model using InstanceNorm. A gated attention mechanism generates two feature representations, which pass through a set of linear and dropout layers for the final predictions. (c) Schematic of a residual block from the Resnet-34-IBN-b model (Right - Adapted from Pan et al.26).
In this work, we use an embedding-level MIL classifier. MIL allows for two types of approaches, instance-level, where classification can be done on each individual component associated with a bag, and embedding-level, where the bag is classified but the meaning of each individual instance is lost. We chose to use the embedding level approach as previous literature has shown that the attention mechanism allows for importance ranking of individual instances within a bag.20 In traditional multiple instance learning, under the collective assumption, all training instances in a given bag contribute equally to the final prediction.19 We modify this, based on previous works, to include a weighted average of instances, which is learned through an attention mechanism. We use the gated attention mechanism proposed previously,20,38 where the weights derived for each patch k is calculated by:
In these equations, represents the specific feature embedding, are the same dimension where has been set to 128, and is the size of the feature embedding, and to generate the weights, which then go through a softmax function which is used to pool feature embeddings.20,38 The idea of an MIL-embedding classifier is that we get a prediction for a set of patches, but do not have to predict each individual patch. We use the attention weights to identify different regions in the tissue that have different weights for prediction.
The final loss function was represented as , where α is set to 0.125 to allow for an equal influence for both prediction tasks based on empirical testing. The model was trained using a single GeForce GTX 1080 GPU for 9 epochs and we chose the model with the lowest validation loss. Supplemental Figure 1 shows the training loss and validation loss as a function of training epochs.
Dataset generation
The slides in the dataset can consist of multiple individual histological slices of the same tissue. To tackle this problem, these slices were grouped into geographical subregions. The subregions were later used to train, validate, and test the predictive models. First, we generated a slide position matrix, consisting of the x and y coordinates of all the tiles per slide. Next, to identify the individual tissue slices, we used density-based spatial clustering of applications with noise (DBSCAN) over the plotted data.28 We used ϵ of 0.3, where ϵ is the maximum distance between two samples for them to be considered in the same neighborhood.28 The clusters identified by the algorithm represent the individual tissue slices in a slide. Finally, the tiles in each cluster were sorted first by x and second by y and were split into subsets of sizes 120 (to prevent memory constraint issues when running the deep learning models). The resulting subsets were used to train, validate, and test the predictive models.
Data analysis and visualization
To visualize the features developed from the MIL-embedding, we used t-distributed stochastic neighbor embedding (tSNE) to reduce the dimensionality of feature vectors extracted from the model.39 We performed dimensionality reduction on both arms of the multi-task learning model, right before the last linear layer, and visualized the purity score and tissue type label by color for each bag based on the slide label. In addition, tissue patches were reconstructed with the predicted label to generate heatmaps for prediction and attention weights (normalized to span from 0 to 1 on a bag level) were used to identify different regions on the slide that were weighted differently by the model. We multiplied the attention weights for the tumor type prediction by the predicted tumor purity, to better identify the tumor regions of interest.
As mentioned, the loss function for purity score prediction generates a probability vector , which is of length K-1, where K is the number of different categories. We generate ROC curves, using the package scikit-learn, based on these probabilities for each of the thresholds set. We average the probability vectors for each slide to get a final probability vector that we use to get the ROC curve and subsequent AUC value. For each cancer type, we calculated F1-score, precision, recall for each class and an overall accuracy score based on the classification_report function in scikit-learn. We aggregate the predictions from all tiles the entire slide and do a majority voting to get slide level tissue type predictions.
We compared the tumor purity score predictions by our model to the predictions generated in a study by Fu et al.24 In this paper, the authors use label smoothing to predict patches for each tile in a fully-supervised approach, where the true label is given as the tumor purity for the sample. For the comparison, we calculate MSE and MAE for both studies based on true versus predicted tumor purity scores of 701 slides that were present in the test datasets of both studies. Our model generates a list of probabilities per tile, where the ith element of the list corresponds to the probability the tile has a tumor purity at least within the ith bin. To calculate a tumor purity score of each tile, we used a probability cutoff of 0.5. Next, we calculated the average tumor purity score of all tiles in a slide to get a tumor purity score on a slide level. Finally, to compare the predictions generated in two studies, we used the MSE and MAE functions from the sklearn.metrics module.
Plots were created in python using matplotlib and seaborn.
Ethics
The study was performed in accordance with relevant guidelines and regulations and was approved by the Institutional Review Board at Weill Cornell Medicine (IRB #1305013903) "Research for Precision Medicine". Informed consent from all participants were obtained. All data was anonymized prior to analysis.
Statistics
Most of our analyses did not require statistics. We report various metrics to assess deep learning model performance, which includes F1-score, precision, recall and AUC-ROC for a TCGA validation set, TCGA test set, and WCM test set. We report median and inter-quartile range for the label distribution of our purity scores for the TCGA dataset.
Role of funders
Funders had no role in study design, data collection, model design, data analyses, interpretation, or writing of the report.
Results
Data and model description
A total of 5390 slides from 3240 patients, comprised of six different cancer types, including adrenal adenocarcinoma (ACC), lung squamous cell carcinoma and lung adenocarcinoma (LUSC & LUAD respectively), invasive breast carcinoma (BRCA), head and neck squamous cell carcinoma (HNSC), prostate adenocarcinoma (PRAD), and low and high grade serous carcinomas (ovarian serous cystadenocarcinoma labeled in TCGA or OV), were used to train, validate, and test our proposed deep learning model (70% | 15% | 15% respectively). The most frequent tumor types were invasive breast carcinoma (BRCA) and lung cancer (LUAD & LUSC), a combination of lung adenocarcinoma and lung squamous cell carcinoma (Supplemental Table 1). The model was tested using two separate cohorts of patients. The TCGA database slides were used for model training and validation (4063 and 921 slides respectively), and a held-out test set (866 slides) from the TCGA database was used to evaluate model performance (TCGA cohort). In addition, we used a Weill Cornell Medicine (WCM) cohort of 78 de-identified H&E slides from 48 patients for evaluating model performance and generalizability (WCM independent cohort).
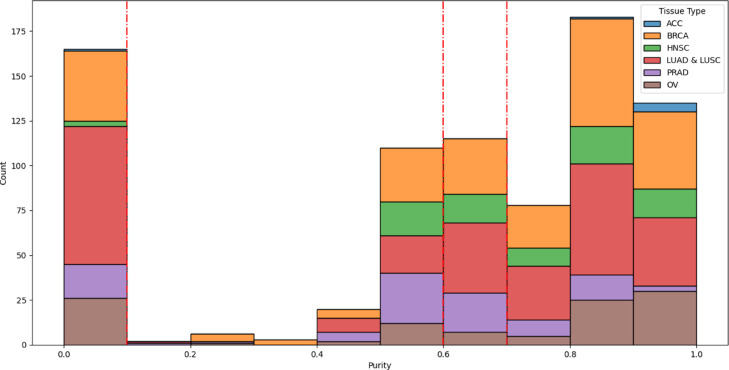
There is variability in the purity distribution between different cancer types. Figure 2 shows the distribution of the held-out TCGA test set purity distribution stratified by tumor type. Here we can see that tumor purity distributions are cancer-type specific, for example OV is skewed towards higher tumor purity, whereas PRAD is skewed to lower tumor purity, when excluding normal tissues. In addition, pathology provided tumor purities overall are skewed towards higher values, 0.7 (0.35-0.85) - median (IQR), for all slides used in this study. In addition, if the normal tissue slides are removed, the values for median (IQR) shift higher to 0.8 (0.67–0.89).
Figure 2.
Representative test set distribution for tumor purity data stratified by tissue type. Red lines show the thresholds used for identifying low vs high tumor purity.
The overall workflow of our model can be seen in Figure 1a. WSIs first get split into a set of tiles, and tiles are then filtered to remove out of focus regions and regions that have little to no tissue present. These patches get passed through a deep learning model, where each original image patch is transformed into a feature representation. These patches are combined using a weighted sum of each feature vector, using an attention mechanism, to obtain two final representations that can be used for two downstream tasks, prediction of tumor purity and prediction of tissue type. Figure 1b shows the schematic for the proposed wsPurity deep learning model. In particular, we used a previously developed Resnet-IBN25 network that has been shown to improve generalizability of deep learning models, especially when there is the presence of color variation, which is common for H&E stained slides. The building block of the Renset-IBN model is shown in Figure 1c and adapted from the previously published manuscript.25
Cancer type prediction
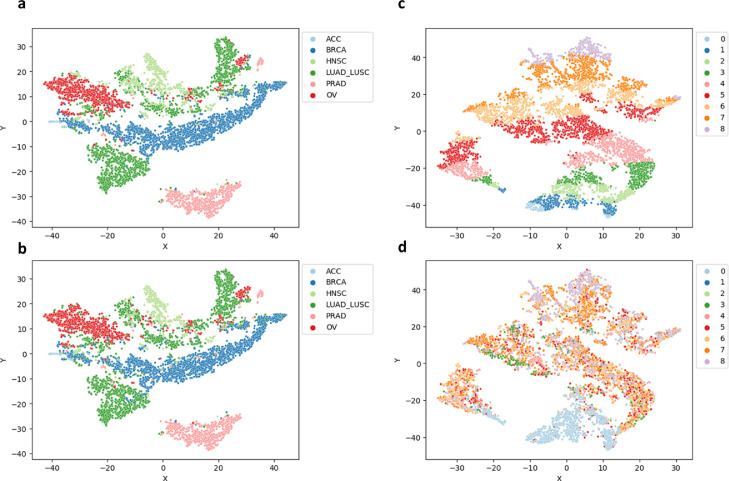
We first aimed to be able to predict cancer type for a given tumor. We generated the labels based on the six cancer type categories (ACC, BRCA, LUSC&LUAD, HNSC, PRAD, and OV) and consider normal solid tissue taken from patients to be grouped into one of these classes as well (we model these patients as having a specific cancer type with 0% tumor purity). As shown in Table 1 our model is capable of predicting cancer type on a held-out set of TCGA patient slides, with an overall accuracy of 93% for both the validation and test sets. The tissue type that performed the worst was ACC, although this cancer type was the most infrequent class with only 17 examples in the validation set and 7 examples in the test set). To visualize how the differences in features extracted from each cancer type, we extract the last fully-connected layer before classification and performed non-linear dimensionality reduction using t-SNE on the feature representation generated per set of 120 patches from a given slide (see Methods for description of slide patching and grouping). From this plot we clearly see separations between the six different cancer types (Figure 4a, b). Interestingly, the features from HNSC and LUAD/LUSC have some mixing with one another (Figure 4). BRCA and PRAD, and OV generally have unique feature representations. Additionally, we do not see any distinct pattern associated with misclassification.
Table 1.
Reported values of F1-score, precision and recall for the tissue type prediction for the validation set, test set (TCGA cohort), and test set (WCM independent cohort).
| Metric | Validation set | Testing set (TCGA) | Testing set (WCM) | |
|---|---|---|---|---|
| ACC | F1-score | 0.84 | 0.77 | - |
| Precision | 0.93 | 0.83 | - | |
| Recall | 0.76 | 0.71 | - | |
| BRCA | F1-score | 0.96 | 0.96 | 0.67 |
| Precision | 0.94 | 0.94 | 1.00 | |
| Recall | 0.99 | 0.97 | 0.50 | |
| HNSC | F1-score | 0.86 | 0.87 | - |
| Precision | 0.80 | 0.79 | - | |
| Recall | 0.93 | 0.96 | - | |
| LUAD and LUSC | F1-score | 0.93 | 0.93 | - |
| Precision | 0.98 | 0.97 | - | |
| Recall | 0.89 | 0.88 | - | |
| OV | F1-score | 0.89 | 0.91 | - |
| Precision | 0.87 | 0.92 | - | |
| Recall | 0.91 | 0.90 | - | |
| PRAD | F1-score | 0.96 | 1.00 | 0.83 |
| Precision | 1.00 | 0.99 | 0.93 | |
| Recall | 0.93 | 1.00 | 0.75 |
Figure 4.
(a, b) tSNE Plots using the feature embedding from the tissue type prediction (compared true vs. predicted, respectively) (c, d) tSNE Plots using the feature embedding from the tumor purity score prediction (compared true vs. predicted, respectively).
When testing tissue type prediction on the WCM cohort, we see that model performance varies depending on cancer type. Overall, the accuracy of the model is 62%, but we see a much higher F1-score for prostate cancer (0.83) compared with breast cancer (0.67). The most common misclassification is breast cancer tissue being misclassified as lung tissue (Supplemental Figure 2). This result may be explained by previous literature, which shows that the spatial patterns are conserved between lung cancer and breast cancer tissue, and specifically that a deep learning model trained on breast cancer can properly make predictions about cancer from lung adenocarcinoma tissue.26 We do see that the most misclassified examples in the TCGA test set for breast cancer slides was also lung cancer (data not shown). In addition, we see that there is a distinction between prostate cancer and breast cancer as the false positive rate for each of these cancers is very low or there are few cases of breast cancer slides predicted as prostate cancer and no prostate cancer slides predicted as breast cancer (precision of 1.0 and 0.93 for breast cancer and prostate cancer respectively). This may indicate that there are distinct features between some cancer types, but for others this prediction may be more difficult to make. In addition, we cannot rule out that some errors may be associated with differences between the tissue preparation and staining between the TCGA cohort and WCM cohort that may influence cancer type prediction.27
Tumor purity prediction
We then look to predict tumor purity, using labels at 10% intervals, in order to provide granularity to predictions, but also allowing for enough examples to be included within each of the categories. These bins were arbitrarily selected and can be altered to suit any other purity based application. To assess how well our model performed, we report both mean squared error (MSE) and mean absolute error (MAE). When looking at model performance between the validation and test sets, we see that model performance for MSE/MAE is 0.0689/0.2079 and 0.0557/0.1867 respectively, indicating good generalizability between unseen TCGA slides. We then compared our model against the fully-supervised approach previously published,24 as well as compared how our model performed in the held-out TCGA cohort compared to the WCM cohort. We identified 701 slides from our test set that overlapped with previously published work in the Fu et al. paper.24 Firstly, we see that our weakly-supervised approach performs better than previous published work, with a MSE/MAE value of 0.0441/0.1659 vs 0 0.1538/0.2967. In addition, we see that our model also generalizes well to our WCM independent test cohort. The performance of tumor purity prediction on the WCM, although significantly smaller in size, is comparable to that of the TCGA held-out data (MSE/MAE - 0.0354/0.1543).
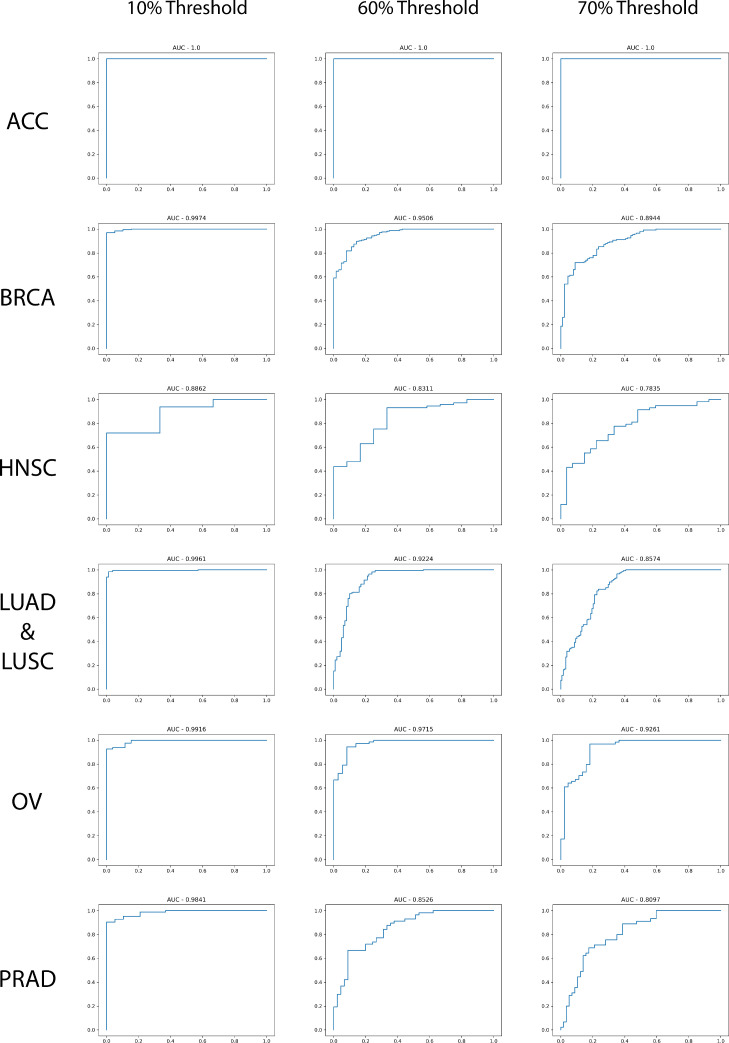
In addition, to validate this model in determining clinically relevant subclasses of tumor purity (high vs. low tumor purity), we generated receiver operating characteristic (ROC) curves based on three separate thresholds for the held out TCGA dataset (Figure 4). The first threshold is for tumor vs. normal tissue prediction. Since we do not have any tumor purity less than 0.09, the 10% threshold can be used to consider the classification between tumor vs. normal groups. What we see is that for the majority of tumor types, the AUC is greater than 0.98. The lowest performing tissue type is HNSC, which may be due to the lower proportion of normal tissue to tumor tissue distribution (∼1:20 normal to tumor ratio). This is much lower than all other cancers besides the most infrequent class, ACC, by a significant amount (the next smallest was PRAD at ∼ 1:5 ratio). ACC performance metrics may not be completely reliable due to the rarity of the class type and the skewed distribution of tumor purities (all values above 80% purity and 1 normal or 0% pure example in the validation and test set), and therefore we caution making direct comparisons for ACC and other tissue types in terms of model performance. Increasing the number of examples or augmenting this dataset may be a useful future endeavor. However, overall, model performance shows an AUC greater than 0.78 for all thresholds and all different cancer types (Figure 3).
Figure 3.
ROC curves of high vs low tumor purity. We set the thresholds at 10% tumor purity, 60% tumor purity, and 70% tumor purity to identify model performance comparing normal vs. tumor tissue.
To visualize how the differences in features extracted from each cancer type for purity prediction, we plotted the features from the last fully connected layer using tSNE similar to the cancer type prediction. When analyzing the feature embeddings for the purity scores, we see that the predicted purity values follow a uniform pattern from highest to lowest. Since these embeddings are on the batch or “bag” level, they may correspond to only a subset of the tissue slice, if the number of tissue patches within a slide exceeded 120, due to computational constraints. Therefore, what we see is that in the extreme cases (i.e. pure tumor or no tumor), the embeddings match well with the predicted tumor type. When the tumor is a mixture of normal and abnormal tissue, we see that there is a combination of many different tumor purities across the slide (Figure 4c, 4d).
Tumor visualization and purity distribution
To assist clinicians and basic scientists, the regions of high tumor purity need to be visualized. In this model, we can identify different regions within the tissue, such as tumor and non-tumor regions (Figure 5). We do this by relying on the attention weights generated from the deep learning model and the output from the purity prediction. Due to the multi-task attention approach, we have two weights per patch per slide, one for tissue type prediction and one for tumor purity prediction. We will focus solely on the tumor purity weights, and compare these regions with pathologist-derived labels of these tumor regions.
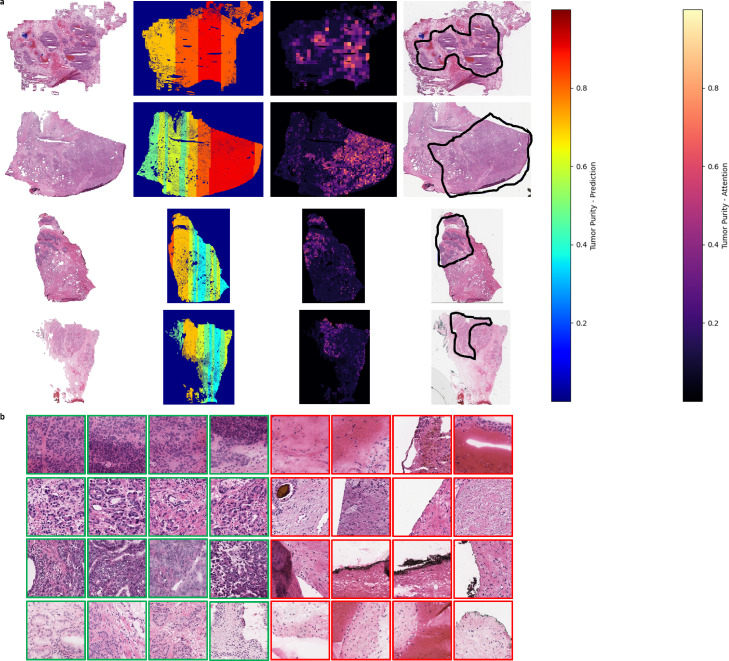
Figure 5.
(a) (Left) A view of the overall tissue architecture. (Middle-Left) The distribution of the tumor purity predictions. (Middle-Right) Attention Maps based on wsPurity. (Right) Pathologist-derived annotations for the tumor region within the slide. (b) Display of the top four (green) and bottom four (red). Rows in A correspond to rows in B. The patches were chosen by normalizing the attention weights using the maximum and minimum values per 120-patch bag, multiplying this normalized attention weight by the predicted purity, and then ranking the values and taking the top four and bottom four patches from this ranked list.
Tissue sections are a priori split into batches of 120 tiles, similar to what has been done in previous multi-task learning methods, as computationally all tiles cannot be analyzed simultaneously.8 We use the x and y coordinates from each tile, to determine the geographical relationship between tiles. We perform density-based spatial clustering (DBSCAN28) on these coordinates, to identify unique tissue sections within the given tissue slide, as multiple sections can be placed on a given slide. We next identify each cluster and sort the x coordinates and then by the y coordinates for each tissue patch. These generate vertical batches that are spatially related and can be passed through the model, as shown by the vertical purity estimates in Figure 5a. Since predictions are done on the batch or “bag” level for embedding classifiers, each 120-patch bag in this process generates a single purity prediction.
As shown in Figure 5a (right-most image column), pathologists annotated what they classified as the tumor regions, for several of the WCM slides, to compare to our attention-based areas of interest for the WCM cohort. The regions of high attention weights correspond well with regions within the tissue that pathologists labeled as tumor. In addition, we can infer higher resolution tumor regions for the tumor purity with respect to regions identified using our model. As shown in Figure 5, we have identified specific regions within a tumor region that have low tumor content (as shown by white arrows on the attention map weights). In addition to the overall tumor location map based on the attention weights, we can identify regions with tumor content within a given slide. We do this by multiplying the predicted purity score by the batch normalized attention weights. Figure 5b shows an example of the top four patches and bottom four patches with this attention weight scoring, where each row of Figure 5b corresponds to the slides in the same row as Figure 5a. We next had a pathologist review these given slides to understand the types of tissue that were identified. Specifically for all examples of the lowest weights, no signs of malignant cells were identified. For each example slide we are able to show examples of cancerous tissue for the given slide. In total, 14/15 patches showed strong evidence for malignant cells. In addition, these patches represent different hallmarks of cancerous tissue. For example, for the first example slide and third example slide the patches show malignant cells that appear to have large sized nuclei compared to the benign or immune cells within the patch. Secondly, in the second and fourth row, we identify small glands as well as poorly developed glands that are indicative of grade 3 prostatic adenocarcinoma. We finally matched these patches to the spatial location within the given slide to confirm that they were within the pathologist drawn regions of interest for tumor regions (Supplemental Figures 5–8).
We also compared how the selection of batches may influence the attention maps. We flipped the sorting of tiles for batch generation, to first sort on the y coordinates and then the x coordinates (horizontal batches). As shown through Supplemental Figure 3, the regions identified do not significantly change based on visual inspection, indicating that our approach is not sensitive to this artificial binning procedure.
We show several different functionalities of our model. First, we show that we can predict the tumor purity, within the entire slide, with a higher accuracy, based on MSE/MAE, as compared to a fully supervised approach, and that we can accurately identify the cancer type within the slide. In addition, we can identify regions within the slide that are tumor positive using attention weights, and we can infer these regions at a higher resolution than what a pathologist would annotate.
Discussion
In this paper, we present a novel weakly-supervised approach to purity estimation from digitized frozen H&E stained slides. Our deep learning-based pipeline extracts several different types of information for pathologists that could benefit or augment their current workflow. Firstly, the model predicts pathologist estimated tumor purity, with accuracy that outperformed a previous approach to purity estimation using fully supervised model training based on a direct comparison of model predictions on the same slides. Secondly, our model outputs a probability of tumor content exceeding a set of predefined thresholds. We show that this approach can then be used to predict high vs low tumor purity for a set of cancer dependent thresholds, which may be useful for predicting clinical outcomes based on previous literature.2 Lastly, our attention mechanism in the model, which aggregates information from different regions of the slide for final purity prediction, can be used for visualization of high and low contributing patches to the final tissue prediction. We show that for this particular task the higher attention weights correspond with regions that are marked as having tumor in the slide based on pathologist-derived estimations of tumor regions. Additionally, these regions are generated at higher resolution than the pathologist annotations, which could be useful in the setting of tumor enrichment for downstream sequencing tasks. Finally, we show that our model can work for many different cancer types, which is supported by previous literature that has shown that different tumor types have conserved spatial patterns.26
While we highlight many attributes of this approach, there are limitations to this work. Firstly, the model does not cover the entire tumor purity spectrum and our training data has skewed purity scores. A small portion, 4% of all slides fell below 50% purity, when not considering normal tissue. While this is the inherent nature of the TCGA database, we see this as a potential area to improve model performance in the very low purity regime. Although our WCM test set had examples of low tumor purity slides (Figure 5), including a more comprehensive assessment of tumor purity, in the low purity regime, is critical for future applications. Our plan could include finding private patient cohorts that have lower purity scores and performing transfer learning or retraining our model on a new set of data. Additionally generative models, such as StyleGAN2 have shown to produce high fidelity images for natural images as well as histological image patches.29 We could use this approach to generate synthetic data, that we can then label to generate artificial aggregated image patches to represent low tumor purity examples. Synthetic data generation may also be helpful as we could assess extreme cases of low tumor purity, where there are very few tumor cells in the entire cohort to find a threshold of predicting whether cancer is present within a tissue to improve our prediction of tumor vs normal slides. Finally, there has been work that shows that formalin-fixed paraffin-embedded (FFPE) slides can be used for RNA-sequencing.30 We can also look to perform transfer learning on a set of FFPE slides, as these slides are used for diagnosis, and these slides maintain a better tissue architecture in most cases compared to frozen sections and are used for diagnostic purposes. Supplemental Figure 4 shows examples of tissue artifacts associated with frozen tissue sections, which are challenging for pathologist assessment of cancer type.
Ideally this approach could have several future applications. Firstly, in cancers such as gliomas, with known clinical outcome correlation with tumor purity, our model can derive a probability estimate of being high or low tumor purity.2 The flexibility of our model to predict this cutoff at different purity values allows for flexibility across different cancer types and can allow for the prediction of whether or not there is cancer within a given slide. Secondly, our approach can assist in precision medicine. Our approach could improve RNA-sequencing results by improving overall tumor content compared to normal tissue during RNA-sequencing of tumors using attention-map guided tumor enrichment. We would need to assess how tumor enrichment would change by comparing our model guided tumor region identification, as compared to pathologist derived tumor region identification, and is something that can be analyzed in future work, potentially within a retrospective study. In addition, our approach could be used as a method to derive a consensus purity estimate, by predicting not only pathologist derived purity estimates, but also incorporating molecular based approaches, such as ABSOLUTE and ESTIMATE, into model predictions. Future work could increase the flexibility of the model to fine-tune model predictions based on the tumor purity information available at a given institution. Another possible direction for this work would be to incorporate more fine-grain information about the different cancer types. One modification would be to include information of specific cancer histological subtypes, molecular based subtypes or mutational status, for which deep learning models have shown promising results, to provide additional information to pathologists for their review.7,31
Data sharing statement
Code for training and the trained models are publicly available on GitHub at https://github.com/ih-lab/wsPurity. TCGA data is available through their website and the WCM data is not publicly available.
Contributors
M.B., P.K., I.H and O.E. conceived of the project. M.B. and V.G. carried out experiments and wrote the manuscript draft. MS, AS, TK, MA and JMM assisted with acquiring and the interpretation of the internal EIPM cohort. MA and JMM provided histopathology interpretation. I.H. supervised the study. All authors read, edited, and approved the final manuscript. M.B., V.G. and I.H. have verified the underlying data.
Declaration of interests
O.E. is scientific advisor and equity holder in Freenome, Owkin, Volastra Therapeutics and OneThree Biotech. The remaining authors declare no competing financial interests.
Acknowledgments
This work was supported by a Starr Cancer Consortium Grant (SCC I15-0027) to I.H. The results shown here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Footnotes
Supplementary material associated with this article can be found in the online version at doi: 10.1016/j.ebiom.2022.104067.
Appendix. Supplementary materials
References
- 1.Mao Y., Feng Q., Zheng P., et al. Low tumor purity is associated with poor prognosis, heavy mutation burden, and intense immune phenotype in colon cancer. Cancer Manag Res. 2018;10:3569–3577. doi: 10.2147/CMAR.S171855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang C., Cheng W., Ren X., et al. Tumor purity as an underlying key factor in glioma. Clin Cancer Res. 2017;23(20):6279–6291. doi: 10.1158/1078-0432.CCR-16-2598. [DOI] [PubMed] [Google Scholar]
- 3.Carter S.L., Cibulskis K., Helman E., et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshihara K., Shahmoradgoli M., Martínez E., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4(1):1–11. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aran D., Sirota M., Butte A.J. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6(1):1–12. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haider S., Tyekucheva S., Prandi D., et al. Systematic assessment of tumor purity and its clinical implications. JCO Precis Oncol. 2020;4:995–1005. doi: 10.1200/PO.20.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woerl A.C., Eckstein M., Geiger J., et al. Deep learning predicts molecular subtype of muscle-invasive bladder cancer from conventional histopathological slides. Eur Urol. 2020;78(2):256–264. doi: 10.1016/j.eururo.2020.04.023. [DOI] [PubMed] [Google Scholar]
- 8.Campanella G., Hanna M.G., Geneslaw L., et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25(8):1301–1309. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahmood F., Borders D., Chen R.J., et al. Deep adversarial training for multi-organ nuclei segmentation in histopathology images. IEEE Trans Med Imaging. 2019;39(11):3257–3267. doi: 10.1109/TMI.2019.2927182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmauch B., Romagnoni A., Pronier E., et al. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat Commun. 2020;11(1):1–15. doi: 10.1038/s41467-020-17678-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kather J.N., Pearson A.T., Halama N., et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25(7):1054–1056. doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma Y., Shrivastava A., Ehsan L., Moskaluk C.A., Syed S., Brown S. Cluster-to-conquer: A framework for end-to-end multi-instance learning for whole slide image classification. Medical Imaging with Deep Learning (MIDL) 2021:682–698. [Google Scholar]
- 13.Pati P., Jaume G., Foncubierta-Rodríguez A., et al. Hierarchical graph representations in digital pathology. Med Image Anal. 2022;75:1–16. doi: 10.1016/j.media.2021.102264. [DOI] [PubMed] [Google Scholar]
- 14.Pinckaers H., Bulten W., van der Laak J., Litjens G. Detection of prostate cancer in whole-slide images through end-to-end training with image-level labels. IEEE Trans Med Imaging. 2021;40(7):1817–1826. doi: 10.1109/TMI.2021.3066295. [DOI] [PubMed] [Google Scholar]
- 15.Lu M.Y., Williamson D.F., Chen T.Y., Chen R.J., Barbieri M., Mahmood F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat Biomed Eng. 2021;5(6):555–570. doi: 10.1038/s41551-020-00682-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bokhorst J.M., Pinckaers H., van Zwam P., Nagtegaal I., van der Laak J., Ciompi F. Learning from sparsely annotated data for semantic segmentation in histopathology images. Medical Imaging with Deep Learning (MIDL) 2019;102:84–91. [Google Scholar]
- 17.Bandi P., Geessink O., Manson Q., et al. From detection of individual metastases to classification of lymph node status at the patient level: the camelyon17 challenge. IEEE Trans Med Imaging. 2018;38(2):550–560. doi: 10.1109/TMI.2018.2867350. [DOI] [PubMed] [Google Scholar]
- 18.Chen C.L., Chen C.C., Yu W.H., et al. An annotation-free whole-slide training approach to pathological classification of lung cancer types using deep learning. Nat Commun. 2021;12(1):1–13. doi: 10.1038/s41467-021-21467-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foulds J., Frank E. A review of multi-instance learning assumptions. Knowl Eng Rev. 2010;25(1):1–25. [Google Scholar]
- 20.Ilse M., Tomczak J., Welling M. Attention-based deep multiple instance learning. ICML. 2018:2127–2136. [Google Scholar]
- 21.Naik N., Madani A., Esteva A., et al. Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains. Nat Commun. 2020;11(1):1–8. doi: 10.1038/s41467-020-19334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomita N., Abdollahi B., Wei J., Ren B., Suriawinata A., Hassanpour S. Attention-based deep neural networks for detection of cancerous and precancerous esophagus tissue on histopathological slides. JAMA Netw Open. 2019;2(11) doi: 10.1001/jamanetworkopen.2019.14645. -e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu M.Y., Chen T.Y., Williamson D.F., et al. AI-based pathology predicts origins for cancers of unknown primary. Nature. 2021;594(7861):106–110. doi: 10.1038/s41586-021-03512-4. [DOI] [PubMed] [Google Scholar]
- 24.Fu Y., Jung A.W., Torne R.V., et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat Cancer. 2020;1(8):800–810. doi: 10.1038/s43018-020-0085-8. [DOI] [PubMed] [Google Scholar]
- 25.Pan X., Luo P., Shi J., Tang X. Two at once: Enhancing learning and generalization capacities via ibn-net. European Conference on Computer Vision (ECCV) 2018:464–479. [Google Scholar]
- 26.Noorbakhsh J., Farahmand S., Namburi S., et al. Deep learning-based cross-classifications reveal conserved spatial behaviors within tumor histological images. Nat Commun. 2020;11(1):1–14. doi: 10.1038/s41467-020-20030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard F.M., Dolezal J., Kochanny S., et al. The impact of site-specific digital histology signatures on deep learning model accuracy and bias. Nat Commun. 2021;12(1):1–13. doi: 10.1038/s41467-021-24698-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ester M., Kriegel H.P., Sander J., Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. KDD. 1996:226–231. [Google Scholar]
- 29.Karras T., Laine S., Aittala M., Hellsten J., Lehtinen J., Aila T. Analyzing and improving the image quality of stylegan. CVPR. 2020:8110–8119. [Google Scholar]
- 30.Pennock N.D., Jindal S., Horton W., et al. RNA-seq from archival FFPE breast cancer samples: molecular pathway fidelity and novel discovery. BMC Med Genom. 2019;12(1):1–18. doi: 10.1186/s12920-019-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coudray N., Ocampo P.S., Sakellaropoulos T., et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24(10):1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodenas P. https://github.com/pedrofrodenas/blur-Detection-Haar-Wavelet. [viewed April 15, 2022 ]
- 33.Tong H., Li M., Zhang H., Zhang C. Blur detection for digital images using wavelet transform. IEEE ICME. 2004;1:17–20. [Google Scholar]
- 34.Paszke A., Gross S., Massa F., et al. Proceedings of the NeurIPS. Vol. 32. 2019. Pytorch: an imperative style, high-performance deep learning library; pp. 1–12. [Google Scholar]
- 35.Ulyanov D, Vedaldi A, Lempitsky V. Instance normalization: The missing ingredient for fast stylization. arXiv preprint arXiv:160708022 2016: 1-6.
- 36.Russakovsky O., Deng J., Su H., et al. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015;115(3):211–252. [Google Scholar]
- 37.Cao W., Mirjalili V., Raschka S. Rank consistent ordinal regression for neural networks with application to age estimation. Pattern Recognit Lett. 2020;140:325–331. [Google Scholar]
- 38.Dauphin Y.N., Fan A., Auli M., Grangier D. Proceedings of the ICML. 2017. Language modeling with gated convolutional networks; pp. 933–941. [Google Scholar]
- 39.Van der Maaten L., Hinton G. Visualizing data using t-SNE. JMLR. 2008;9(11):2579–2605. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.