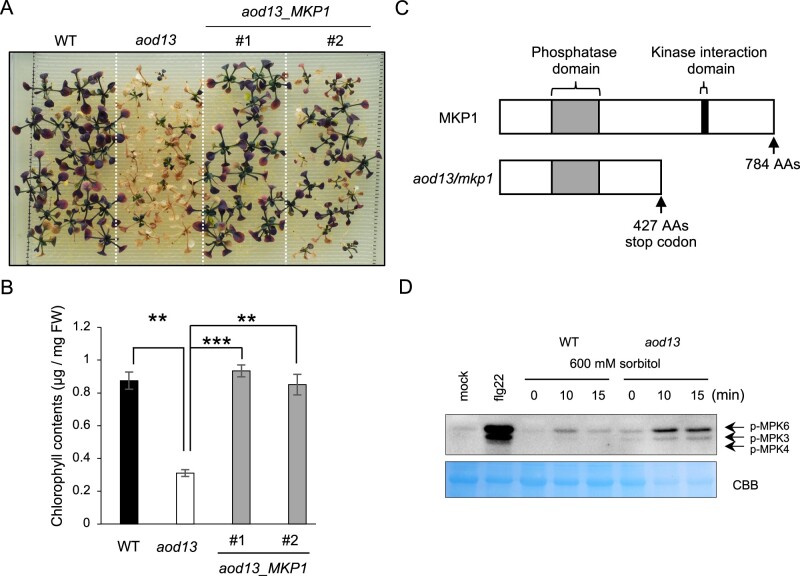
Figure 3.
Identification of the causal gene in aod13. A, Complementation test performed by transforming aod13 with MKP1. T3 homozygous plants transformed with native promoter: MKP1 (aod13_MKP1) derived from Bu-5 WT were used in an acquired osmotolerance assay. B, Chlorophyll contents of WT, aod13, and aod13_MKP1 as described in (A). Differences between Bu-5 WT and aod13, and between aod13 and aod13_MKP1 were analyzed by Student’s t test (mean ± se, n = 8, **P < 0.01, ***P < 0.001). C, Schematic representation of MKP1 and aod13 (i.e. mkp1) proteins. AAs, amino acids. D, Seven-day-old Bu-5 WT and aod13 plants were treated with or without 600 mM sorbitol for the times indicated. As a positive control for activated MPK3/6 detection, the plants were treated with flg22, a conserved 22-amino-acid peptide derived from P. syringae flagellin, or water (mock). Proteins from treated leaf tissue were separated by SDS–PAGE and immunoblotting was performed using an anti-Phospho-p44/p42 MAPK (anti-pTEpY) (upper panel) to detect phosphorylated MAPKs. Extracted total proteins (input) stained with Coomassie brilliant blue served as a loading control (bottom panel).