Figure 4.
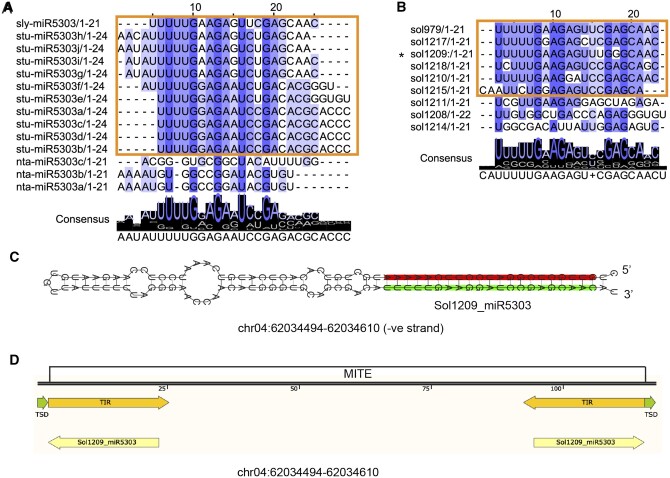
miR5303 aligns to MITE TIR sequences. The previously annotated Solanaceae-specific miRNA family, miR5303, exhibits many features that disqualify it as a bonafide miRNA. The majority of the miR5303 miRBase entries include mature sequences that are 24 nt in length, indicating that these sRNAs are likely derived from siRNA producing loci (A). miR5303 miRBase entries are highly variable in their sequence; we identified a consensus sequence to isolate true miR5303 family members (highlighted in a box and shown in Supplemental Figure S3) (A). miRNAs >22-nt in length were filtered out; however, the predicted miR5303 family members that are 21 or 22 nt follow the same 5′-consensus sequence (UUUUG) as the miRBase entries (B). The predicted precursors of miR5303 family members have near-perfect hairpin folding, indicating that these loci are likely inverted repeats, rather than real miRNA precursors (C), and BLAST alignment of miR5303 family members to the plant MITE database produces a perfect alignment between the miR5303 species and MITE TIR sequences (D).