Fig. 7.
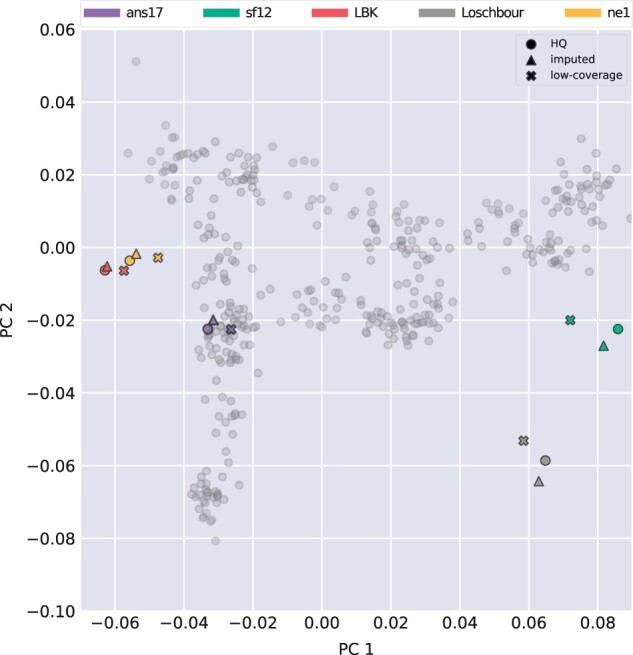
PCA comparing HQ, imputed, and low-coverage data for the five evaluation individuals. A reference PCA was defined based on genotypes of modern European samples from the Human Origins (Patterson et al. 2012) data set, after which the KDR method was used to estimate scores of ancient individuals. Modern individuals are indicated by gray dots and ancient individuals colored according to the legend. Not all modern individuals are visible in the plot, see Supplementary Fig. 1 for a view of the full data set. The low-coverage data points correspond to the genotypes that have been downsampled to 1×, filtered, and used as input to imputation, which was performed using configuration 3 (Table 1), with a posterior filter of minimum genotype probability of 0.99 applied.
