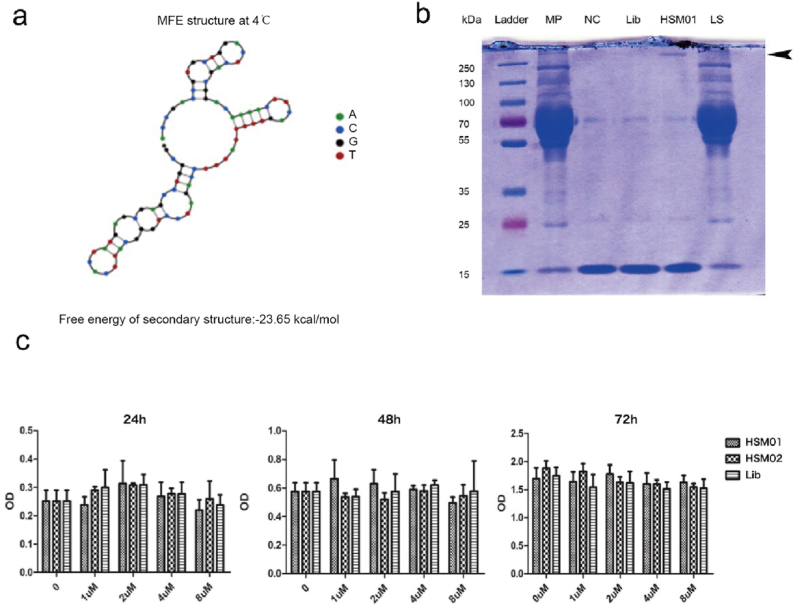
Fig. 5.
Aptamer HSM01 structure prediction, protein precipitation, and cell treatment experiments. a. Nupack software (http://www.nupack.org/) was used to predict the secondary structure (2D structure) of aptamers. b. the HSKMC membrane protein was incubated with biotin-conjugated HSM01, Lib, or blank, respectively, and then the biotin affinity magnetic beads were added to elute the target protein complex. Then, the HSM01 sample, Lib sample, and NC sample were collected. The remaining supernatant of the HSM01 sample was the last supernatant (LS sample). The above samples (MP sample, HSM01 sample, Lib sample, NC sample, and LS sample) were analyzed through SDS-PAGE electrophoresis and Coomassie Brilliant Blue Staining. A very specific protein band in the HSM01 lane was found above 250 kd. The band and control were sent for LC-MS/MS QSTAR analysis (Supplementary Table 3 and 4). c. The treatment of HSM01 and HSM02 on HSKMCs showed the effect of the aptamer on cell proliferation in 24 h, 48 h, and 72 h, respectively.