Fig. 1.
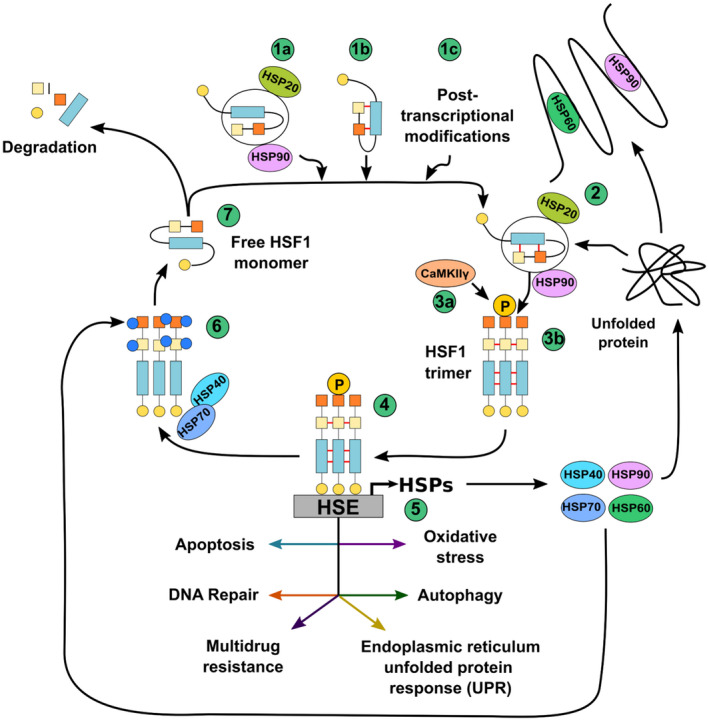
HSF1 activation. HSF1 is usually present in the cell in an inactivated form. Inactivation of HSF1 occurs mainly by three mechanisms: (1a) HSP90 binding to HSF1, (1b) HSF1 stabilization through the formation of a leucine zipper structure within the protein (red lines), or (1c) through post‐transcriptional modifications, such as acetylation, sumoylation, and phosphorylation. (2) HSF1 is activated when an increase in misfolded proteins occurs in the cell, such as after heat shock (increased environmental temperature). HSF1 activation involves the release of monomeric HSF1 from chaperones, such as HSP20 and HSP90 (3a). Once activated, HSF1 monomers interact together to form a trimer that is stabilized by leucine zippers (red lines) (3b) and is phosphorylated by the calcium/calmodulin‐dependent protein kinase II gamma (CaMKIIγ). (4) HSPs act as molecular chaperones for the correct folding of numerous proteins in the cell. (5) HSF1 binds to DNA sequences in the genome, namely heat shock elements (HSE) in the promoters of genes encoding for heat shock responses, such as heat shock proteins (HSPs) promoting their transcription. HSF1 also promotes the transcription of genes involved in the regulation of apoptosis, DNA repair, modulation of drug resistance, unfolded protein response (UPR) at the endoplasmic reticulum, autophagy, and oxidative stress, among others. (6) Acetylation (blue circles) of HSF1 at Lys80 destabilizes its interaction with the DNA. HSP40 together with HSP70 bind to specific sites in HSF1 monomers leading to a destabilization of the trimer. (7) Excess HSF1 is degraded through the SCFβ‐TrCP pathway, and only a basal amount of inactive HSF1 remains in the cell.
