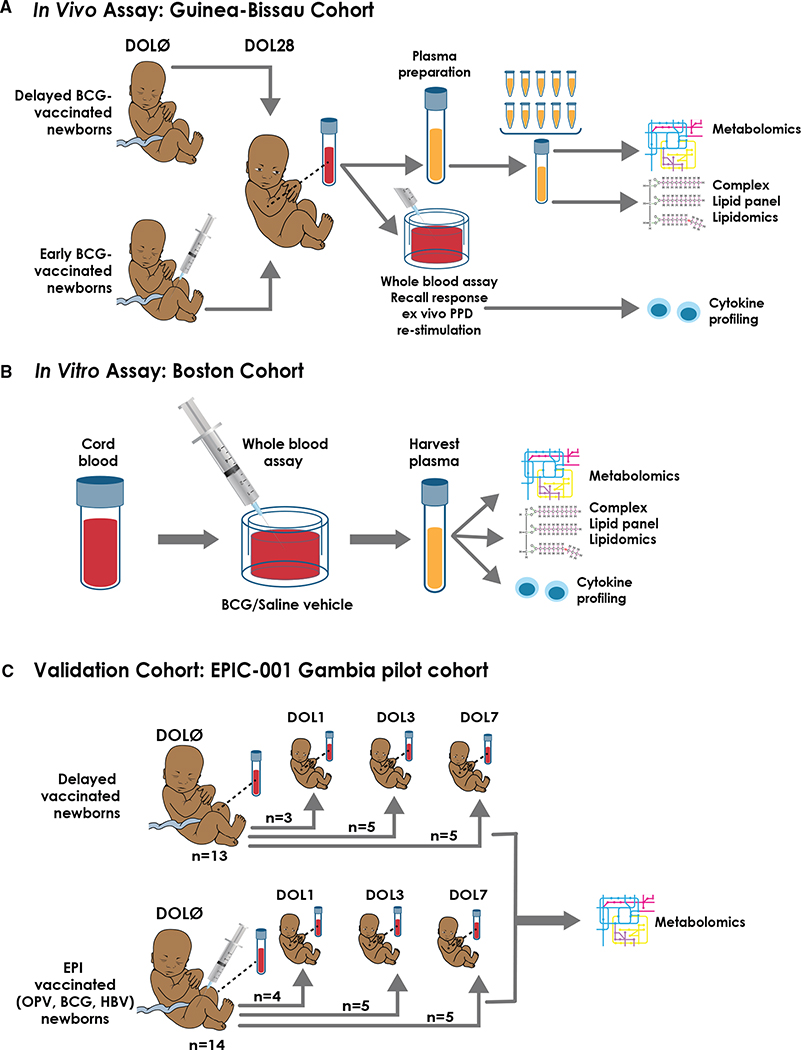
Figure 1. Schematic of in vivo, in vitro, and validation cohort for BCG vaccination experimental setup.
(A) Study design and participant plasma biosample flow chart. Low-birthweight newborns in Guinea-Bissau were assigned to delayed BCG (catch-up BCG after blood collection) or early BCG (vaccinated with BCG at birth). In an immunological study nested within the trial, capillary blood samples were collected 4 weeks after randomization to investigate the effect of BCG on TLR-agonist-induced and PPD antigen recall cytokine responses. Plasma biosamples of sufficient volume were utilized for subsequent metabolomic and lipidomic assays. To ensure sufficient sample volume for robust metabolite detection, metabolomics and complex lipid panel lipidomics were conducted on pooled plasma from n = 10 per sample for newborns of the same sex and vaccine treatment.
(B) To further assess BCG’s impact on the plasma metabolome, we modeled BCG vaccine responses in vitro in cord blood from a cohort of newborns in Boston, USA. Similarly, supernatants from these in vitro stimulated samples were subjected to high-throughput metabolomic, lipidomic, and cytokine/chemokine assays, and results indexed to the same participant’s vehicle (saline) control.
(C) For validation purposes, we compared the metabolomics profile of Guinea-Bissau in vivo and Boston in vitro cohorts with another independent newborn group from The Gambia (West Africa) (Lee et al., 2019). Newborns were assigned to either receive the Expanded Program on Immunization (EPI) vaccines (OPV, BCG, and HBV) at birth or delayed during the first week of life to study the effect of vaccine responses on early life immune ontogeny. Paired samples were analyzed by indexing their DOL0 (day of birth) samples and a follow-up time point at DOL1, DOL3, or DOL7. Regardless of their time point assignment, the newborns were combined into their assigned treatment groups (EPI-vaccinated versus delayed-vaccinated) for analysis.