Abstract
Objective
The liver is the primary internal metabolic organ that coordinates whole body energy homeostasis in response to feeding and fasting. Genetic ablation or pharmacological inhibition of calcium/calmodulin-dependent protein kinase kinase 2 (CaMKK2) has been shown to significantly improve hepatic health and peripheral insulin sensitivity upon overnutrition with high fat diet. However, the precise molecular underpinnings that explain this metabolic protection have remained largely undefined.
Methods
To characterize the role of CaMKK2 in hepatic metabolism, we developed and challenged liver-specific CaMKK2 knockout (CaMKK2LKO) mice with high fat diet and performed glucose and insulin tolerance tests to evaluate peripheral insulin sensitivity. We used a combination of RNA-Sequencing, glucose and fatty acid istotopic tracer studies, a newly developed Seahorse assay for measuring the oxidative capacity of purified peroxisomes, and a degenerate peptide libarary to identify putative CaMKK2 substrates that mechanistically explain the protective effects of hepatic CaMKK2 ablation.
Results
Consistent with previous findings, we show that hepatic CaMKK2 ablation significantly improves indices of peripheral insulin sensitivity. Mechanistically, we found that CaMKK2 phosphorylates and regulates GAPDH to promote glucose metabolism and PEX3 to blunt peroxisomal fatty acid catabolism in the liver.
Conclusion
CaMKK2 is a central metabolic fuel sensor in the liver that significantly contributes to whole body systems metabolism.
Keywords: Insulin resistance, Glucose metabolism, Fatty acid metabolism, Peroxisome, Kinase signaling
Abbreviations: CaMKK2, Calcium/Calmodulin Dependent Protein Kinase Kinase 2; NAFLD, Non-alcoholic fatty liver disease; HCC, Hepatocellular carcinoma; NASH, Non-alcoholic steatohepatitis; T2DM, Type 2 Diabetes Mellitus; HFD, High fat diet; GTT, Glucose tolerance test; ITT, Insulin tolerance test; OCR, Oxygen consumption rate; HCD, High carbohydrate diet; CLAMS, Comprehensive Lab Animal Monitoring System; PTT, Pyruvate tolerance test; RER, Respiratory Exchange Ratio; ECAR, Extracellular acidification rate
Highlights
-
•
Hepatic CaMKK2 is sufficient to toggle peripheral insulin sensitivity.
-
•
CaMKK2 is required for hepatic glucose metabolism.
-
•
Hepatic CaMKK2 balances glycolytic and fatty acid metabolism.
-
•
CaMKK2 promotes glycolysis through regulation of GAPDH.
-
•
CaMKK2 suppresses fatty acid metabolism through Pex3.
1. Introduction
The balance of circulating glucose, controlled by peripheral sensitivity to insulin, is influenced by a variety of factors including environment, genetics, and diet. Chronic glucose intolerance promotes the onset of metabolic disease that often includes dyslipidemia, hyperglycemia, and peripheral adiposity [1]. Acutely, the body is well-equipped to store surplus calories in subcutaneous fat depots [2]. However, chronic overnutrition promotes aberrant accumulation of lipid in the liver, leading to the onset of non-alcoholic fatty liver disease (NAFLD) [[2], [3], [4]]. Prolonged exposure to excess hepatic lipid compromises normal liver metabolic function, promotes peripheral insulin resistance and the onset of Type 2 Diabetes Mellitus (T2DM) [5]. While NAFLD responds to changes in diet and caloric expenditure, unresolved inflammation brought on by excess lipid progresses to steatohepatitis (NASH) and ultimately hepatocellular carcinoma (HCC) [[6], [7], [8]]. Steatohepatitis currently represents the leading cause of liver transplants and is expected to worsen as cases of NAFLD continue to rise [9,10]. Thus, there is a critical need to define causal relationships between dietary caloric stress and disruption of peripheral glucose homeostasis to identify therapeutic targets to improve metabolic liver disease outcomes.
The liver serves as the central metabolic hub for the body, functioning in concert with other metabolic tissues to maintain glucose levels in accordance with caloric intake [11]. Under homeostatic conditions, the liver primarily utilizes fatty acids for energy production and supplies glucose to other peripheral tissues, most notably the brain [11,12]. In response to prolonged fasting, the liver is unique in its ability to manufacture glucose de novo from non-carbohydrate precursors [11]. These vital hepatic metabolic functions are compromised during chronic overnutrition, blunting fatty acid catabolism as result of increased lipid burden in the liver [13,14]. Our laboratories have identified calcium/calmodulin-dependent protein kinase kinase 2 (CaMKK2) as a critical rheostat for the regulation of hepatic glucose and lipid metabolism [[15], [16], [17]]. Congenic ablation of CaMKK2 in the context of high-carbohydrate diet feeding compromises insulin action and increases hepatic steatosis [16]. Conversely, loss of CaMKK2 promotes insulin sensitivity and reduces hepatic steatosis in mice challenged with high-fat diet (HFD) [15]. In line with these findings, pharmacological inhibition of CaMKK2 reversed major hallmarks of NAFLD in wild-type mice fed HFD [17]. Taken together, these findings support CaMKK2 as an important molecular regulator that balances hepatic lipid and glucose metabolism in response to dietary challenge.
Within hepatocytes, energy production from glucose and lipids requires the coordinated action of multiple organelle systems [13]. To meet the energetic demands of the cell, mitochondria rely on communication with the endoplasmic reticulum, nucleus, and peroxisomes [18,19]. Glycolytic byproducts are imported into mitochondria along with lipids for β-oxidation to fund ATP production [20]. While mitochondria account for the majority of fatty acid β-oxidation, peroxisomes also contribute to lipid metabolism [21]. Although peroxisomes primarily degrade damaging reactive oxygen species (ROS), they also perform essential functions for processing very long chain fatty acids for transport into the mitochondria [22]. Also, peroxisomes can sustain mitochondrial β-oxidation in the context of genetic or structural damage [[23], [24], [25]]. In such instances, peroxisomes oxidize fatty acids to shorter chain lengths that freely diffuse across the mitochondrial membrane by circumventing the carnitine transport system [26,27]. Furthermore, peroxisomes have been reported to be the only organelle that directly interacts with lipid droplets, highlighting their importance to fatty acid metabolism [28,29]. Given the phenotypic improvements to liver health observed upon genetic loss or pharmacological inhibition of CaMKK2 [[15], [16], [17]], we sought to mechanistically evaluate how this central metabolic kinase functions as a fuel source selector to regulate the hepatic and peripheral responses to overnutrition.
2. Results
2.1. Hepatic CaMKK2 regulates glucose tolerance and peripheral insulin sensitivity
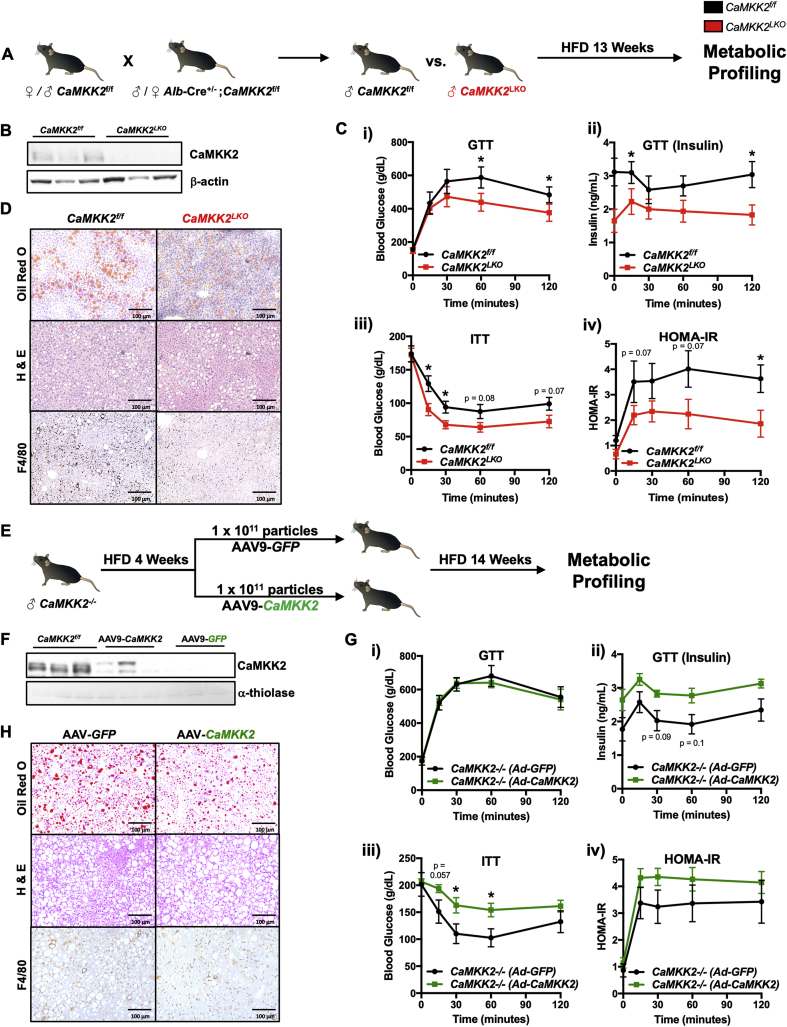
As acute loss of hepatic CaMKK2 has been shown to improve insulin sensitivity [15], we sought to validate these findings and explore its mechanism of action. To investigate the function of CaMKK2 in hepatic metabolism, we generated liver-specific knockout mice (CaMKK2LKO) using a breeding scheme that yielded CaMKK2LKO and wild-type (CaMKK2f/f) littermates (Figure 1A). CaMKK2f/f and CaMKK2LKO male mice were placed on HFD for 13 weeks and subjected to metabolic profiling prior to euthanasia. As expected, immunoblot analysis confirmed CaMKK2 protein expression was lost in the livers of CaMKK2LKO mice, but retained at normal expression levels in the brain and spleen (Figure 1B and Figs. S1A). CaMKK2LKO mice showed no protection from obesity on HFD, which is anticipated given that the protection observed in whole-body CaMKK2 knockout mice primarily results from reduced signaling through neuropeptide Y (NPY) (Fig. S1Bi) [30]. Furthermore, we found no differences in fat or lean mass in the absence of hepatic CaMKK2 (Figs. S1Bii-iii). Ad libitum blood glucose levels remained comparable between WT and CaMKK2LKO mice throughout the experiment, a phenotype that is distinct from that of the CaMKK2 whole body knockout (Fig S1C). However, under fasting/refed conditions, CaMKK2LKO mice showed improvements in glucose homeostasis, suggesting that loss of hepatic CaMKK2 may influence glycolytic metabolism (Fig. S1D).
Figure 1.
CaMKK2 regulates peripheral glucose tolerance and insulin sensitivity. (A) Schematic overview of the breeding strategy and experimental approach for metabolic profiling in response to HFD feeding. Briefly, C57BL/6J CaMKK2f/f and CaMKK2LKO male mice were aged to 6 months and placed on HFD for 13 weeks. Mice were then subjected to CLAMS, GTT, ITT, and PTT. Following metabolic profiling, mice were euthanized, and livers isolated for downstream phenotypic and molecular analysis. (B) Immunoblot of CaMKK2 and β-actin (loading control) from livers of CaMKK2f/f and CaMKK2LKO mice. (C)CaMKK2LKO mice show protection against markers of insulin resistance. (i) Measurement of blood glucose levels in response to a glucose challenge (glucose tolerance test, GTT) in CaMKK2f/f (WT) (black line; n = 5) and CaMKK2LKO mice (red line; n = 7). (ii) Insulin levels during GTT of CaMKK2f/f (black line; n = 5) and CaMKK2LKO mice (red line; n = 7). (iii) Blood glucose levels during an insulin tolerance test (ITT) in CaMKK2f/f (black line; n = 5) and CaMKK2LKO mice (red line; n = 7). (iv) HOMA-IR of CaMKK2f/f (black line; n = 5) and CaMKK2LKO mice (red line; n = 6) calculated as: fasting insulin (microU/l) x fasting glucose (nmol/l)/22.5. (D) Histology of liver sections from CaMKK2f/f and CaMKK2LKO mice stained for Oil Red O, H & E, and F4/80. (E) Schematic overview of the in vivo approach for assessing glycemia upon hepatic rescue of CaMKK2 expression. Briefly, 5-month-old C57BL/6J male CaMKK2−/− mice were acclimated to HFD for 4 weeks. Mice were retro-orbitally injected with 1 × 1011 particles of AAV9-GFP (n = 6 mice) or AAV9-CaMKK2 (n = 7 mice) and maintained on HFD for 14 weeks. Mice were subjected to a GTT and ITT prior to euthanasia. (F) Immunoblot of CaMKK2 and α-thiolase (loading control) in livers from CaMKK2f/f, AAV-CaMKK2, and AAV-GFP injected mice. (G) AAV-CaMKK2 injected mice show susceptibility to markers of insulin resistance. (i) Measurement of blood glucose levels by GTT in AAV9-GFP (black line; n = 6) and AAV-CaMKK2 injected mice (green line; n = 7) and. (ii) Insulin levels during GTT of AAV9-GFP (black line; n = 6) and AAV-CaMKK2 injected mice (green line; n = 7). (iii) Blood glucose levels during ITT of AAV9-GFP (black line; n = 6) and AAV-CaMKK2 injected mice (green line; n = 7). (iv) HOMA-IR of AAV9-GFP (black line; n = 6) and AAV-CaMKK2 injected mice (green line; n = 7). (H) Histology of liver sections from AAV9-GFP and AAV9-CaMKK2 injected mice for Oil Red O, H & E, and F4/80. Data are represented as the mean ± s.e.m. ∗P < 0.05.
When subjected to a glucose tolerance test (GTT), CaMKK2LKO mice displayed lower blood glucose accompanied by decreased insulin (Figure 1Ci-ii), indicating improved glucose tolerance and peripheral insulin sensitivity. Similarly, CaMKK2LKO mice maintained lower blood glucose during an insulin tolerance test (ITT) and had a lower HOMA-IR, indicating improved peripheral insulin sensitivity (Figure 1Ciii-iv). Consistent with previously published data, CaMKK2LKO mice showed a trend toward decreased blood glucose when subjected to a pyruvate tolerance test (PTT) (Fig. S1Ei), suggesting a potential defect in gluconeogenesis (Fig. S1Eii) [15]. Collectively, these findings are consistent with acute loss of hepatic CaMKK2 [15] and further support the notion that hepatic ablation of CaMKK2 improves whole-body insulin sensitivity. Histological analysis revealed no differences in gross morphological changes (H & E), neutral lipid staining (Oil Red O) or macrophage infiltration (F4/80) between WT and CaMKK2LKO mice (Figure 1D). Furthermore, assessment of whole-body energy expenditure using the Comprehensive Lab Animal Monitoring System (CLAMS) revealed no differences in VCO2, VO2, or the respiratory exchange ratio (RER) (Figs. S1F). Serum ALT and AST were quantified as indicators of hepatic health and found to be similar between the two genotypes (Figs. S1Gi-ii). Finally, serum triglycerides, glycerol, and free fatty acids were also similar in both genotypes (Figs. S1Giii-vi).
As acute or stable deletion of hepatic CaMKK2 was sufficient to markedly improve peripheral insulin sensitivity, we next evaluated the impact of re-expression of hepatic CaMKK2 on glycemic control. CaMKK2−/− mice were acclimated to HFD for four weeks. This acclimation period was selected based on the opposing phenotypes observed in CaMKK2−/− mice fed HFD [16]. Following four weeks of HFD feeding, CaMKK2f/f and CaMKK2LKO mice were administered AAV9-GFP or AAV9-CaMKK2 via retroorbital injection and were maintained on HFD for an additional 14-weeks post-injection (Figure 1E). Immunoblot analysis confirmed successful rescue of hepatic CaMKK2 expression in mice injected with AAV9-CaMKK2, although at slightly lower levels than wild-type (Figure 1F). This observed reduction in CaMKK2 expression in AAV9-CaMKK2 injected mice was expected given that exogenous gene expression is maximal approximately five weeks post-adenoviral injection [31].
In response to rescue of hepatic CaMKK2 expression, we observed no changes in body weight or ad libitum blood glucose levels (data not shown). Leveraging the exact GTT conditions used in our CaMKK2LKO experiment (Figure 1Ci), mice with re-expressed hepatic CaMKK2 maintained similar blood glucose levels compared with control mice (Figure 1Gi). Quantitation of insulin levels following the GTT revealed that AAV9-CaMKK2 treated mice maintained nearly identical glycemia when compared to control treated mice but with significantly higher insulin, suggesting a decrease in insulin sensitivity (Figure 1Gii). Consistent with these findings, AAV9-CaMKK2 injected mice displayed higher blood glucose during an ITT, again indicative of decreased insulin sensitivity (Figure 1Giii). Although HOMA-IR showed no significant difference, AAV9-CaMKK2 injected mice showed a decrease in insulin action (Figure 1Giv). Consistent with results from CaMKK2LKO mice (Figure 1D), histological analyses to examine hepatic fat content and macrophage infiltration revealed minimal differences between AAV9-CaMKK2 and control treated mice (Figure 1H). Taken together, these data collectively support the conclusion that hepatic CaMKK2 is a major determinant in the development of insulin resistance in response to overnutrition with HFD.
2.2. CaMKK2 reprograms hepatic metabolism
To interrogate the underlying mechanism by which CaMKK2 facilitates metabolic reprogramming of the liver, we performed a comprehensive RNA-Seq analysis from livers of CaMKK2LKO and control mice in response to HFD feeding (Figure 2A). Ontological pathway analysis revealed that hepatic CaMKK2 ablation upregulated several pathways associated with lipid metabolism, including fatty acid and bile acid metabolism, peroxisomes, and oxidative phosphorylation, with a concurrent decrease in glycolysis and inflammatory pathways (Figure 2B). Next, we validated candidate gene expression from the RNA-Seq using qPCR to confirm the increased expression of lipid metabolism and peroxisomal gene programs, and a concomitant decrease in inflammatory targets (Figure 2C). Supporting these findings, Scd1 was the most upregulated gene associated with lipid metabolism, as it is required for long-chain fatty acid synthesis. Interestingly, while our ontological analysis highlighted a decrease in genes associated with glycolytic metabolism, validation of these targets showed a surprising increase that may be due to compensatory effects. Further analysis of the RNA-Seq data revealed that the metabolic reprogramming conferred by loss of hepatic CaMKK2 results in a healthier gene expression profile (Figure 2D). We compared our CaMKK2LKO RNA-Seq data with gene expression datasets from human patients with healthy obesity, NAFLD, NASH and HCC using Gene Set Enrichment Analysis (GSEA). We found a positive correlation between gene signatures from our CaMKK2LKO mice and those from patients with healthy obesity (Figure 2Di). Moreover, loss of hepatic CaMKK2 increased the expression of genes normally downregulated in NAFLD, along with a decrease in genes associated with NASH and HCC (Figure 2Dii-iv). Normalized enrichment scores support these conclusions, as we found positive enrichment only with the healthy obese gene set, but negative enrichment for the NAFLD, NASH, and HCC gene sets (Figure 2Dv). Consistent with previous reports from our laboratory, the gene signature produced by hepatic CaMKK2 ablation correlated with better overall and disease-free survival in HCC patients (Figure 2Ei-ii) [32]. Taken together, these data highlight that although the CaMKK2LKO mice show no protection from obesity, the hepatic transcriptional landscape resulting from loss of hepatic CaMKK2 suggests protection from the full spectrum of metabolic liver disease, likely due to improvements in lipid metabolism (Figure 2B–C).
Figure 2.
CaMKK2 reprograms hepatic metabolism. (A) Livers isolated from CaMKK2f/f and CaMKK2LKO mice from Figure 1A were used to assess changes in gene expression by RNA-Seq. Heatmap representation of statistically significant genes changed relative to CaMKK2f/f at ≥ 1.5X cutoff. (B) Significant gene signatures from (A) were used to identify ontological gene programs impacted by hepatic loss of CaMKK2. (C) Quantitative real-time PCR analysis of genes involved in lipid metabolism, glucose metabolism, and inflammation on RNA isolated from livers of CaMKK2f/f (black bars; n = 3) and CaMKK2LKO mice (red bars; n = 6). (D) Gene Set Enrichment Analysis (GSEA) comparing the CaMKK2LKO gene signature with gene expression datasets from (i) healthy obese patients, (ii) NAFLD patients, (iii) NASH patients and (iv) HCC patients, which are stratified from high (red) to low (blue) expression. (v) Bar graph representation of normalized enrichment scores of GSEA calculated from (i-iv), respectively. (E) Kaplan–Meier (i) survival and (ii) disease-free survival plots of HCC patients (n = 242) comparing a positive correlation with the CaMKK2LKO gene signature (red lines) or negative correlation (blue lines) with the CaMKK2LKO gene signature. Data are represented as the mean ± s.e.m. Unpaired Student's t-test was used to determine statistical significance. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
To evaluate the transcriptional effects in response to rescue of hepatic CaMMK2 expression, we performed an RNA-Seq analysis from livers of mice injected with either AAV-GFP or AAV-CaMKK2 (Fig S2A). While rescue of hepatic CaMKK2 differentially impacted the transcriptional landscape, we found a reduction in the number of statistically altered genes compared to the RNA-Seq performed in CaMKK2LKO mice and, as such, fewer pathways showed significant enrichment during analysis. Interestingly, the top upregulated ontological pathway in response to AAV9-CaMKK2 treatment was glycolysis/gluconeogenesis, complementing the results obtained from our CaMKK2LKO RNA-Seq (Fig. S2B). Due largely to the lower number of altered genes, GSEA failed to reach statistical significance, but we noted a trend toward negative enrichment for genes associated with oxidative phosphorylation in the AAV9-CaMKK2 treated group, further supporting the observation that loss of hepatic CaMKK2 appears to sponsor an increase in lipid metabolism, consistent with our previous report [15] (Fig. S2C).
2.3. Hepatic CaMKK2 ablation negatively impacts cell autonomous glucose metabolism
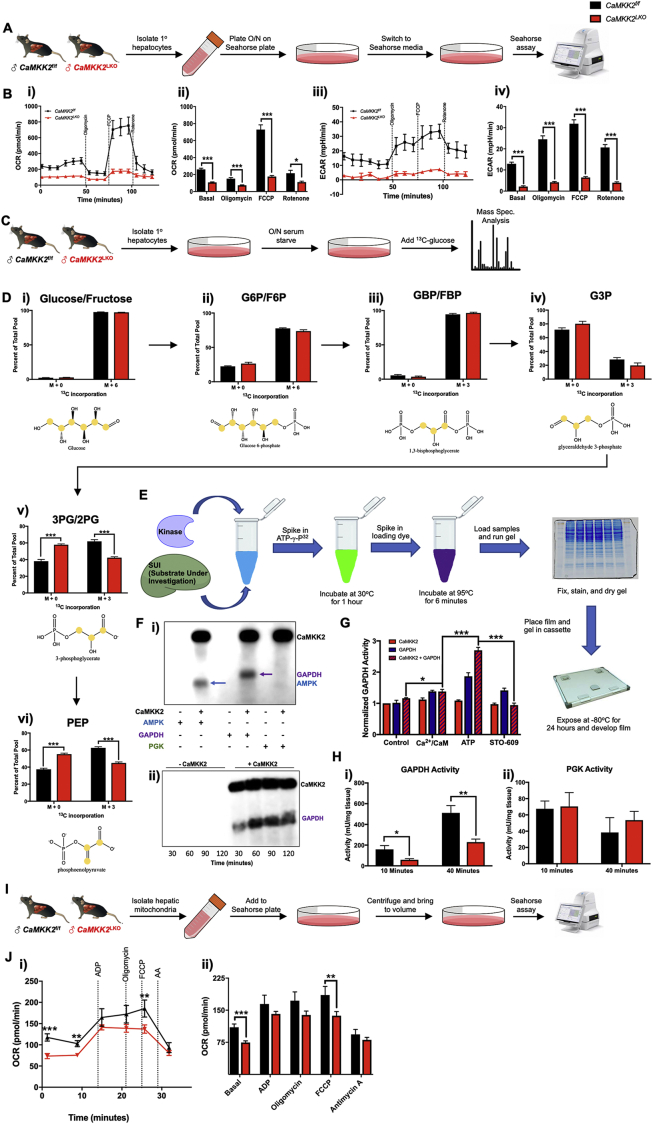
Acute loss of hepatic CaMKK2 is sufficient to improve glycemic control, at least in part through the activation of fatty acid metabolic programs [15]. Consistent with these findings, ontological pathway analysis of our RNA-Seq data indicate that stable genetic ablation of hepatic CaMKK2 confers a metabolic shift towards lipid utilization, perhaps at the expense of glycolysis (Figure 2B). Leveraging the current gold standard for cell autonomous metabolic activity, the Seahorse assay was used to determine the effect of CaMKK2 on mitochondrial respiration. Primary hepatocytes isolated from CaMKK2f/f and CaMKK2LKO mice were plated overnight and subjected to the MitoStress assay the following day (Figure 3A). Hepatocytes isolated from CaMKK2f/f (WT) mice showed higher oxygen consumption rates (OCR), indicative of improved aerobic activity (basal OCR levels) and capacity (OCR levels after FCCP) (Figure 3Bi-ii). Similarly, we found that CaMKK2f/f hepatocytes displayed higher extracellular acidification rates (ECAR), which is an index of improved glycolytic metabolism (Figure 3Biii-iv). This conclusion is further supported by the OCR/ECAR ratio, which is an indicator of the preferred metabolic fuel source (Fig. S3Ai-ii). Higher OCR/ECAR ratios often reflect elevated lipid utilization, while a lower ratio suggests a propensity towards glucose metabolism. The OCR/ECAR ratio was extremely low in the CaMKK2f/f hepatocytes, providing additional evidence for their reliance on glucose. Conversely, the OCR/ECAR ratio in CaMKK2LKO hepatocytes remained high, suggesting that genetic loss of hepatic CaMKK2 alters the ability of these cells to metabolize lipids at the expense of glucose.
Figure 3.
Hepatic CaMKK2 ablation negatively impacts cell autonomous glucose metabolism. (A) Schematic for Seahorse assay using primary hepatocytes. Primary hepatocytes were isolated from CaMKK2f/f and CaMKK2LKO male mice fed normal chow. Hepatocytes were plated for 16 h, and mitochondrial respiration assessed with the Seahorse Mito Stress Assay. (B) Mito Stress Seahorse assay results: (i) Oxygen consumption rate (OCR) of primary hepatocytes isolated from CaMKK2f/f (black line) and CaMKK2LKO (red line) mice. (ii) Extracellular acidification rate (ECAR) of primary hepatocytes isolated from CaMKK2f/f (black line) and CaMKK2LKO (red line) mice. (C) Schematic for stable isotope tracing using 13C6-glucose. Primary hepatocytes from CaMKK2f/f and CaMKK2LKO mice were isolated and plated in normal growth media. Hepatocytes were starved overnight (∼16 h) in glucose-free DMEM, after which 10% FBS and 12 mM 13C6-glucose or 12C6-glucose (control) was added to the media and incubated for 72 h. (D) Incorporation of 13C into glycolytic intermediates. Location of 13C incorporation and measurement as percent of total pool in: (i) glucose/fructose, (ii) glucose-6-phosphate/fructose-6-phosphate, (iii) glucose-(1,6)-bisphosphate/fructose-(1,6)-bisphosphate, (iv) glyceraldehype-3-phosphate, (v) 3-phophoglycerate/2-phosphoglycerate, and (vi) phosphoenolpyruvate. (E) Schematic of the in vitro kinase assay for validation of putative kinase targets. Recombinantly purified human CaMKK2 protein and potential targets were incubated in the presence of ATP-γ-32P at 30 °C for 1 h. (F) (i)In vitro kinase assay of CaMKK2 with AMPK1-312 (positive control, lane 2) or putative targets (GAPDH (lane 4) and PGK (lane 6)). Presence of a band indicates incorporation of 32P by CaMKK2. (ii) Time-course in vitro kinase assay comparing phosphorylation levels of GAPDH at different timepoints in the absence (lanes 1–4) or presence (lanes 5–8) of CaMKK2. (G) Measurement of GAPDH activity in CaMKK2f/f (black bars) and CaMKK2LKO (red bars) livers isolated from (1A) after 10 min and 30 min, respectively. (H) Schematic for measurement of mitochondrial respiration in isolated mitochondria. Mitochondria were isolated from livers of CaMKK2f/f and CaMKK2LKO mice and assayed according to Sakamuri et al. [78]. (I) (i) Oxygen consumption rate of isolated mitochondria from CaMKK2f/f (red line) and CaMKK2LKO (black line) mouse livers. (ii) Oxygen consumption from (i) graphed as average of averages. Data are represented as the mean ± s.e.m. Unpaired Student's t-test was used to determine statistical significance. One-way ANOVA was used to analyze mitochondrial respiration. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
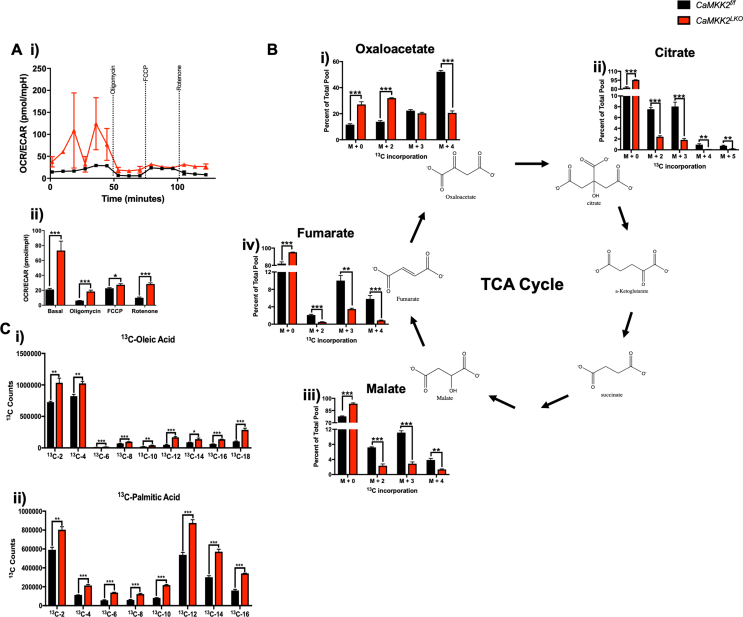
We considered two plausible explanations that could account for the significantly lower OCR values observed in CaMKK2LKO hepatocytes: 1) glucose may not be effectively transported into the cell; or 2) there is a defect in glucose metabolism. To resolve these two possibilities, we performed stable isotope tracing with 13C6-glucose in hepatocytes isolated from CaMKK2f/f and CaMKK2LKO mice (Figure 3C). Importantly, we identified that hepatocytes from both genotypes readily uptake labeled glucose at the same rate, as indicated by the equivalent amounts of 13C6-glucose incorporation (Figure 4Di). These tracer studies also revealed similar glycolytic rates between both genotypes through the formation of the first three metabolic intermediates of glucose metabolism (Figure 4Dii-iv). We found a significant difference in the conversion of G3P to 3PG/2 PG in the absence of hepatic CaMKK2 that was maintained through the formation of PEP (Figure 3Dv-vi). Although labeled glycolytic intermediates constituted a much smaller percent of the total pool, this deficit was maintained throughout the TCA cycle (Figs. S3Bi-iv). Next, we repeated a second stable isotope tracer experiment with 13C6-glucose at 72-hours to determine the fate of the glycolytic carbon and found that CaMKK2LKO hepatocytes produced significantly more labeled long-chain fatty acids, notably oleic and palmitic acid (Figs. S3Ci-ii).
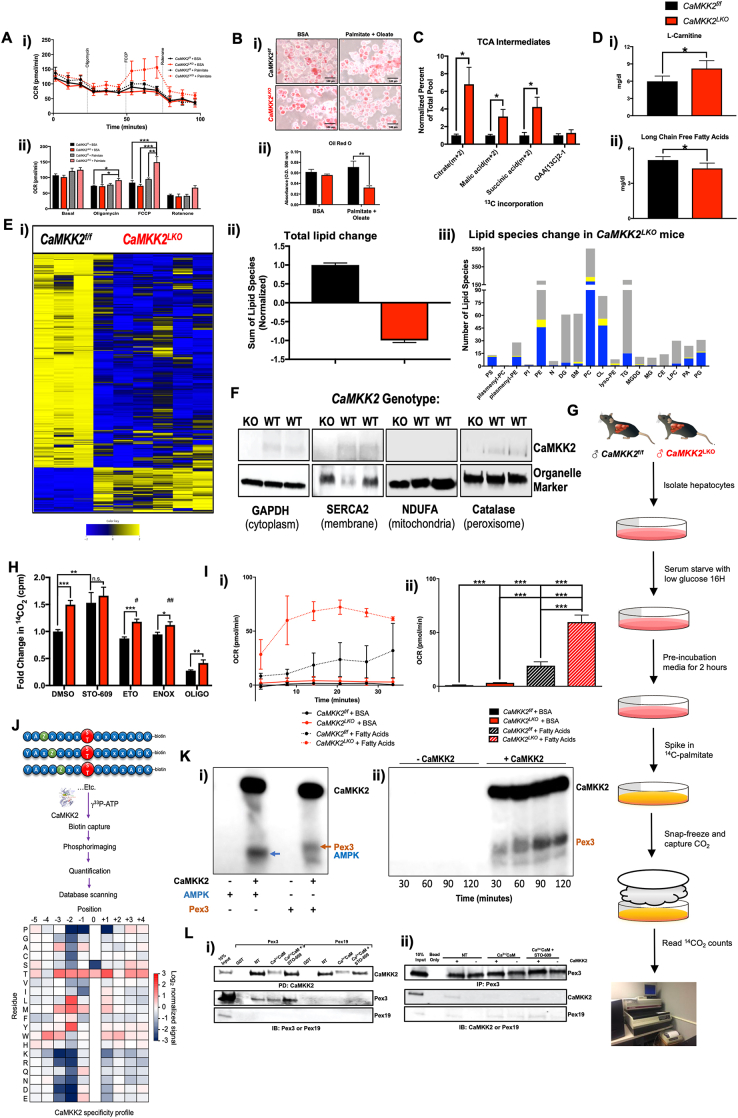
Figure 4.
Hepatic CaMKK2 ablation increases cell autonomous fatty acid metabolism. (A) (i) OCR of primary hepatocytes isolated from CaMKK2f/f (black lines) and CaMKK2LKO (red lines) mice in the presence (dotted lines) or absence (solid lines) of palmitate as measured by Seahorse technology and (ii) average at each stage represented by bar graphs. (B) (i) Primary hepatocytes from (A) were stained with Oil Red O following the Seahorse assay, photographed (ii) and the dye extracted, and absorbance measured at O.D. 500 nm. (C) Primary hepatocytes isolated from CaMKK2f/f and CaMKK2LKO mice were serum starved, followed by addition of FBS and 13C-palmitate or unlabeled palmitate (control) and incubated for 6 h 13C incorporation into TCA cycle intermediates in CaMKK2f/f (black bars) and CaMKK2LKO (red bars) was measured and represented as percent of the total pool normalized to CaMKK2f/f incorporation. (D) Livers from CaMKK2f/f (black bars) and CaMKK2LKO (red bars) were assayed for levels of (i)l-carnitine and (ii) long chain free fatty acids. (E) Lipidomic analysis of livers from CaMKK2f/f and CaMKK2LKO mice: (i) Heatmap representation of the change in lipid species and (ii) quantification of total lipid landscape in livers from CaMKK2f/f (black bars) and CaMKK2LKO (red bars) mice. (iii) Bar graph representing the number and direction of lipid species changes in CaMKK2LKO relative to CaMKK2f/f livers. Blue denotes decreased lipid, yellow denotes increased lipid, and grey denotes lipids that remained unchanged. PE phamenyl-PC (plasmenyl-glycerophophocholines); plasmenyl-PE (plasmenyl-gycerophophoethanolamines); PI (glycerophosphoinositols); PE (gylcerophosphoethanolamines); N (sphinganines); DG (diacylgycerolipids); SM (sphingomyelins); PC (glycerophosphocholines); CL (cardiolipins); lyso-PE (lyso-glycerophosphoethanolamines); TG (triacylgycerolipids); MGDG (monoacylgycerolipids/diacylgycerolipids); MG (monoacylgycerolipids); CE (cholesterol esters); LPC (lyso-glycerophophocholines); PA (glycerophosphates); PG (glycerophosphogycerols). (F) Immunoblot analysis of hepatic CaMKK2 protein expression in the cytoplasmic, membrane, mitochondrial, and peroxisomal fractions. (G) Schematic of β-oxidation assay from primary hepatocytes isolated from CaMKK2f/f and CaMKK2LKO mice, serum starved O/N, incubated in pre-incubation media, and treated with 14C-palmitate. (H) Results from the β-oxidation represented as fold change in 14CO2 counts between CaMKK2f/f (black bars) and CaMKK2LKO (red bars) mice after treatment with DMSO, 20 mM STO-609 (CaMKK2 inhibitor), 20 mM Etomoxir (ETO; mitochondrial Cpt1 inhibitor), 150 mM enoximone (ENOX; peroxisomal phosphodiesterase inhibitor), or 10 mM oligomycin (OLIGO; electron transport chain inhibitor, negative control). #: denotes statistical significance compared to DMSO-treated primary hepatocytes from CaMKK2LKO mice. (I) Peroxisomes isolated from CaMKK2f/f and CaMKK2LKO livers were assayed by Seahorse: (i) OCR of peroxisomes isolated from CaMKK2f/f (black lines) and CaMKK2LKO (red lines) mice in the presence of BSA (solid lines) or fatty acids (dotted lines). (ii) Average of averages of OCR readings from (i). (J) Schematic of degenerate peptide library analysis used to identify putative CaMKK2 phospho-targets and the actual residues identified. (K) (i)In vitro kinase assay with recombinantly purified CaMKK2 and Pex3. AMPK1-312; positive control. (ii) Time-course in vitro kinase assay comparing phosphorylation levels of Pex3 at different timepoints in the absence (lanes 1–4) or presence (lanes 5–8) of CaMKK2. (L) (i) Immunoblot analysis following pull-down of GST-CaMKK2 in the presence of Pex3 or Pex19 under control conditions (NT), activated conditions (Ca2+/CaM), and inhibited conditions (Ca2+/CaM + STO-609). (ii) Immunoblot analysis of CaMKK2, Pex3 and Pex19 following immunoprecipitation of Pex3. Data are represented as the mean ± s.e.m. Unpaired Student's t-test was used to determine statistical significance. One-way ANOVA was used to analyze peroxisomal respiration ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Our 13C-glucose tracer experiment not only highlighted a defect in glycolysis, but also exposed putative points of regulatory control by CaMKK2. Conversion of glyceraldehyde 3-phosphate (G3P) to 3-phosphogylcerate (3-PG) is a two-step process. First, glyceraldehyde phosphate dehydrogenase (GAPDH) catalyzes the production of 1,3-bisphosphoglycerate (1,3-BPG) from 3-PG. Next, phosphoglycerate kinase (PGK) converts 1,3-BPG to 3-PG [33]. To determine whether GAPDH or PGK are phospho-targets of CaMKK2, we performed an in vitro kinase assay (Figure 3E), and found only GAPDH to be directly phosphorylated by CaMKK2 (Figure 3Fi). Additionally, we found a time-dependent increase in GAPDH phosphorylation in the presence of activated CaMKK2 (Figure 3Fii). We further evaluated GAPDH as an effector of CaMKK2 action by assessing the functional impact of phosphorylation by CaMKK2 on GAPDH activity. Using purified proteins, we found that GAPDH activity is significantly enhanced in the presence of fully activated CaMKK2 (Figure 3G). Consistent with these findings, in vivo GAPDH activity is markedly attenuated in liver extracts of CaMKK2LKO mice (Figure 3Hi). Importantly, the activity of PGK remained unaffected by loss of hepatic CaMKK2 as expected since it could not be directly phosphorylated by CaMKK2 (Figure 3Hii). Taken together, these data demonstrate that CaMKK2 phosphorylation of GAPDH functions as an activation mark that bolsters GAPDH activity and subsequently promotes glucose catabolism.
Since hepatic ablation of CaMKK2 conferred defects in glycolytic metabolism by blunting the enzymatic activity of GAPDH, we hypothesized that this blockade could be rescued with pyruvate to bypass the apparent defect in glycolysis. We isolated mitochondria from livers of CaMKK2f/f and CaMKK2LKO mice for analysis in the Seahorse assay (Figure 3I) and assessed their oxidative potential when administered pyruvate. While loss of CaMKK2 significantly reduced basal OCR levels, the magnitude of this effect was less than when glucose was used as the sole fuel source (Figure 3Ji-ii). Interestingly, mitochondria isolated from CaMKK2LKO livers failed to show an increase in OCR following the addition of the proton uncoupler, FCCP (Figure 3B). This lack of response to FCCP suggests that CaMKK2LKO hepatocytes are functioning at near maximum capacity for ATP production. While much of the oxidative advantage of CaMKK2f/f hepatocytes is abrogated when pyruvate is used instead of glucose as the primary fuel source, these findings suggest that CaMKK2 function likely intersects with additional downstream metabolic steps that have yet to be fully characterized.
2.4. Hepatic CaMKK2 ablation increases cell autonomous fatty acid metabolism
Since loss of hepatic CaMKK2 sponsors an increase in the expression of fatty acid gene programs (Figure 2B), combined with the observation that long-term 13C6-glucose tracing revealed an increase in long-chain fatty acids (Figs. S3Ci-ii), we next examined the effect of lipid substrates on the oxidative capacity of hepatocytes in the absence of CaMKK2. To this end, we performed a β-oxidation Seahorse assay wherein the primary fuel source was a 2:1 mixture of oleic:palmitic acids to mimic the ratio of these long chain fatty acids in our in vivo HFD studies (Figure 1A,E). Although basal respiration was nearly identical across all treatment groups, a noticeable difference was found upon addition of FCCP. Hepatocytes isolated from CaMKK2LKO mice showed significant improvements in OCR (Figure 4Ai-ii). These results are indicative of not only their proclivity to utilize lipids but suggest that cells devoid of CaMKK2 may be more dependent on fat metabolism as their primary fuel source. Hepatocytes from the β-oxidation Seahorse assay (Figure 4A) were subjected to Oil Red O (ORO) staining to visualize neutral lipids. The lipid content of hepatocytes from both genotypes was similar when treated with BSA as a control (Figure 4Bi-ii), consistent with our histological findings from whole liver (Figure 1D). However, upon treatment with oleic and palmitic acid, CaMKK2LKO hepatocytes showed a significant reduction in ORO staining compared to WT cells. These findings are highly consistent with our phenotypic data and provide a plausible cell autonomous explanation for why mice devoid of hepatic CaMKK2 show whole body metabolic improvements when challenged with excess dietary fat.
As CaMKK2LKO hepatocytes metabolize lipid more effectively than glucose, we next performed stable isotope experiments using 13C-palmitate to monitor the oxidative fate of fatty acid carbon (Figure 3C). We found that CaMKK2LKO hepatocytes contained significantly higher levels of 13C-labeled TCA intermediates (Figure 4C), which differs from our findings using 13C6-glucose (Fig. S3B) and provides further support that CaMKK2 ablation confers an increased reliance on lipid metabolism. Moreover, we identified an increase in l-carnitine while long-chain free fatty acids were decreased in serum from CaMKK2LKO mice fed HFD (Figure 4Di-ii). These results are both positive indices of increased lipid metabolism. To evaluate the impact of hepatic CaMKK2 on the broader lipid landscape at the organ level, lipidomic analysis of whole liver extracts revealed that loss of CaMKK2 induced significant changes in lipid content and composition (Figure 4Ei). Our lipidomic analysis was focused primarily on phospholipid species and we found a marked reduction of these lipids in CaMKK2LKO livers compared to WT (Figure 4Eii & Figs. S4A). Next, we analyzed the impact of hepatic CaMKK2 ablation on individual lipid species (Figure 4Eiii), exposing decreased levels of phosphatidylethanolamines (PE) and phosphatidylcholines (PC) that have been implicated as biomarkers of metabolic and inflammatory disease [[34], [35], [36]]. We also found that loss of hepatic CaMKK2 significantly increased levels of unsaturated lipid species in the liver (Fig. S4B). Furthermore, there was an increase in carnitine-associated metabolites in the livers of CaMKK2LKO mice, pointing toward increased lipid metabolism (Fig. S4C).
Considering that loss of hepatic CaMKK2 significantly impacts fuel source selection, metabolism and the overarching lipid landscape of the liver, we next sought to identify the subcellular localization of CaMKK2. We employed differential centrifugation techniques aimed at isolating and purifying cellular organelles to homogeneity. Initially, liver lysates were fractionated to isolate the cytoplasm, nucleus, and plasma membrane. In line with previous findings, we confirmed CaMKK2 expression in the cytoplasm and identified its association with the plasma membrane fraction (Figure 4F). Conversely, no CaMKK2 was found in the hepatic nuclear fraction (Fig. S4D). Next, we isolated individual organelles from whole liver to further evaluate the subcellular localization of CaMKK2. Perhaps unexpectedly, the mitochondrial fraction was devoid of CaMKK2 (Figure 4F). Similarly, the lysosomal, Golgi, and endoplasmic reticulum fractions contained no detectable CaMKK2 (Fig. S4D). However, CaMKK2 was clearly present in the peroxisomal fraction (Figure 4F), which is consistent with the ontological analysis of our RNA-Seq data (Figure 2B). Peroxisomes generally function as detox centers for the cell, as they breakdown and dispose of reactive oxygen species (ROS) [21,37]. Peroxisomes also play a vital role in lipid pre-processing of very long chain fatty acids to shorter chain species that more readily traverse the mitochondrial membrane for β-oxidation and ATP production [18,27,38]. This latter function is vital to human health as peroxisomal disorders, such as Zellweger Syndrome, result from the accumulation of aberrant fatty acids rather than ROS damage [39]. To facilitate this lipid processing function, peroxisomes express a unique set of β-oxidation enzymes that metabolize fatty acids into simpler substrates, including succinate [38,40,41]. For this reason, peroxisomal engagement can compensate for reduced mitochondrial output, as often occurs in the context of diabetes and metabolic syndrome [24,39,42,43].
Since CaMKK2 localizes to peroxisomes, we evaluated the contribution of peroxisomal engagement to β-oxidation of fatty acids by using the conventional β-oxidation assay with 14C-palmitate as the primary fuel source and captured 14CO2 is a proxy for lipid metabolism (Figure 4G). CaMKK2LKO hepatocytes showed a 50% increase in β-oxidation under basal conditions (Figure 4H). Treatment of CaMKK2f/f (WT) hepatocytes with STO-609, a CaMKK2 inhibitor, increased β-oxidation levels but not in hepatocytes isolated from CaMKK2−/− mice (Figure 4H). Furthermore, CaMKK2−/− hepatocytes showed reduced levels of β-oxidation when treated with etomoxir and enoximone, selective inhibitors of mitochondrial and peroxisomal β-oxidation, respectively [44]. This decrease in lipid oxidation was not observed in WT (CaMKK2f/f) hepatocytes, suggesting a lower dependency on lipid metabolism. Given that the reduction in β-oxidation following enoximone treatment mirrored the reductive effects of etomoxir, we reasoned that the increase in oxidative capacity likely reflects higher peroxisomal engagement that, in turn, leads to improved mitochondrial respiration. Using newly developed methodology from our laboratory [45], we isolated viable peroxisomes for use in the Seahorse assay. In the absence of any fuel source, peroxisomes isolated from WT and CaMKK2−/− livers displayed similar OCR outputs (Figure 4Ii-ii). Addition of palmitic acid stimulated OCR output in both genotypes but peroxisomes isolated from CaMKK2LKO showed nearly a two-fold increase in OCR relative to WT (Figure 4Ii-ii). These findings clearly highlight the metabolic contributions of peroxisomes to fatty acid oxidation and emphasize their functional importance to the development of metabolic disease.
To identify putative peroxisomal effectors of CaMKK2 action, we conducted a degenerate peptide library screen to define the optimal consensus motif for CaMKK2 phosphorylation (Figure 4J). The resulting consensus motif was queried against the human proteome to identify putative CaMKK2 phospho-targets. Our bioinformatic search identified over 500 unique proteins and an ontological analysis of these proteins showed enrichment of pathways associated with lipid metabolism and transport (Fig. S4E). More specifically, this screen highlighted peroxisomal biogenesis factor 3 (Pex3) as a putative target of CaMKK2 and an in vitro kinase assay validated that Pex3 is directly phosphorylated by activated CaMKK2 (Figure 3, Figure 4Ki). Consistent with an enzyme–substrate mode of action, CaMKK2-dependent phosphorylation of Pex3 increased in a time-dependent manner (Figure 4Kii). Pex3 is a peroxisomal membrane protein essential for peroxisomal maturation [[46], [47], [48], [49]]. Metabolically, Pex3 is associated with regulation of lipid droplet size via its interaction with Pex19 [49,50]. Given that lipid droplets are a source of fat within the cell, we reasoned that CaMKK2 phosphorylation of Pex3 might alter its interaction with Pex19 by preventing their association, thus resulting in production of larger lipid droplets. Importantly, we found no differences in Pex3/Pex19 protein expression in the absence of hepatic CaMKK2 (Fig. S4F). Using a pull-down of CaMKK2, we demonstrate that CaMKK2 selectively and directly interacts with Pex3 but not Pex19 (Figure 4Li). Additionally, we show that Pex3 and Pex19 directly interact and that CaMKK2 enhances this interaction, indicating that CaMKK2 perhaps functions to sequester this protein complex, thereby suppressing peroxisomal engagement and blunting fatty acid oxidation (Figure 4Lii). Immunofluorescence staining for Pex3/Pex19 revealed a modest increase in their co-localization in CaMKK2LKO livers compared to WT, suggesting that phosphorylation of Pex3 by CaMKK2 may indeed function to prevent its interaction with Pex19 as a means of regulating fatty acid metabolism (Figure S4G).
In conclusion, our findings support that CaMKK2 acts as a fuel source selector in hepatocytes. Our data demonstrate that CaMKK2 phosphorylates GAPDH to promote glycolysis, and Pex3 to inhibit lipid metabolism via decreased peroxisomal engagement. These results not only support CaMKK2 as a viable therapeutic target in the context of metabolic disease, but also highlight the importance of peroxisomal lipid oxidation in the context of obesity and demonstrate how their increased recruitment may encourage or restore normal hepatic metabolic function.
3. Methods
3.1. Primary hepatocyte isolation
Mouse primary hepatocytes were isolated from CaMKK2f/f and CaMKK2LKO male littermate mice, pooled and plated in triplicate as previously described [15]. Briefly, mice were anesthetized with isoflurane until under a surgical plane and maintained on isoflurane for the entire isolation procedure. The liver was perfused with PBS (w/o Mg2+ or Ca2+) containing 1% HEPES (v/v) and 0.019% EGTA (w/v) at 37 °C via cannulation of the inferior vena cava (IVC) and egress through the portal vein at a rate of 5.5 ml/min. Following PBS perfusion, the liver was perfused with PBS (with Mg2+ and Ca2+) containing 0.052% collagenase (w/v) at a rate of 5.5 ml/min for approximately 4 min. Following collagenase digestion, the liver was surgically excised, placed in a sterile petri dish, minced with scissors, and incubated with William's E media containing 10% FBS (v/v), 1% glutamine/gentamycin (v/v), 0.55% pen/strep (v/v), 0.0009% insulin-transferrin-sodium selenite (ITS) (w/v), 0.00036% glucagon (v/v) for approximately 3 min. The resulting mixture was first passed through a sterile 40 mesh stainless steel filter, followed by a 60-mesh filter, and washed with an additional 20 ml of plating media. The resulting suspension was centrifuged at 50×g for 2 min at 4 °C with no brake. The supernate was decanted, and the resulting pellet resuspended in 25 ml of plating media followed by the addition of 12 ml of Percoll (GE) and centrifuged again at 50×g for 10 min at 4 °C with no brake. The supernate was discarded and the resulting cell pellet containing purified primary hepatocytes was resuspended in 10 ml of plating media and cells were counted on a Countess 2 cell counter per the manufacturer's recommendations.
3.2. AAV generation
The AAV9-CaMKK2 plasmid was created using a previously generated CaMKK2 plasmid [51]. Restriction enzyme sites were added to the 5′ and 3′ ends of the full-length gene for insertion into the AAV9 vector backbone. The 5′ primer (NdeI GGTACATATGATGGATTACAAAGACGATGACGATAAGGGAGGCTCATCATGTGTCTCTAGC) and 3′ primer (Sal1 - CGTCGTCGACTTACTCGGGCTCCATGGCCTCCTCC) were engineered to PCR clone the human full-length CaMKK2 cDNA using Eco-Plus Polymerase per manufacturer's instructions, with annealing temperature set to 74 °C and an annealing time of 2 min. Max Efficiency™ DH5α competent E. coli (Thermo, 18258012) were transformed with the AAV-CAG-GFP vector (AddGene #37825) per the manufacturer's instructions. A single colony was isolated, and the plasmid DNA was purified using the PureLink™ HiPure Plasmid Maxiprep Kit (Invitrogen, K210006) per the manufacturer's instructions. An aliquot of the vector was diluted to a final concentration of 1 μg/μl. The vector and purified PCR product were subjected to double restriction enzyme digestion (NdeI and SalI, New England Biosciences) in 3.1buffer per the manufacturer's protocol. Reactions were separated on a 1% agarose gel and products were gel purified using GeneClean III Kit (MP Bio, 111001-600). A T4 ligation reaction was carried out using 50 μg of vector and 37.5 μg of insert (3:1 vector:insert size). Products were separated on a 1% agarose gel to evaluate successful ligation. Positive clones were excised, gel purified, and confirmed by DNA sequencing. The final product was submitted to the gene vector core at Baylor College of Medicine, along with the AAV-CAG-GFP vector (negative control) for AAV9 generation.
3.3. Mice (breeding, genotyping, general phenotyping)
All mouse experiments were performed in accordance with the Animal Care Research Committee at Baylor College of Medicine. We confirmed that a single copy of Cre was required to delete CaMKK2 from the liver via immunoblotting, and thus the described breeding scheme was used. Alb-Cre+/-; CaMKK2f/f mice were generated by crossing CaMKK2f/f mice with Alb-Cre+/+ mice to generate Alb-Cre-/+; CaMKK2f/+. The Alb-Cre-/+; CaMKK2f/+ mice were crossed with CaMKK2f/f mice. Genotyping with gene- and Cre-specific primers were used to identify progeny that were Alb-Cre+/-; CaMKK2f/f, or CaMKK2LKO, and these mice were used as breeders to establish the colony. All mice were bred and maintained on a pure C57BL6/J background. Mice were maintained in a temperature controlled (23°C) facility with a 12-hr light/dark cycle. Mice were fed High Fat (Bioserve #F3282) mouse diet ad libitum with free access to food and water.
3.4. Histology (ORO, H&E)
Fresh liver tissue was fixed in 4% paraformaldehyde for 48 h and embedded in paraffin. Hematoxylin and eosin (H&E) staining was used to evaluate gross morphology, and Sirius Red was used to examine fibrosis. An antibody specific to F4/80 (murine macrophage marker, Thermo Fisher Scientific, Cat#MF48000, 1:100) was used to visualize macrophage infiltration. Frozen-block preparation was performed by embedding liver tissues in OCT compound. Oil Red O staining was used to visualize neutral lipid accumulation as previously described [17]. All staining was carried out by the pathology core at Baylor College of Medicine.
3.5. Immunofluorescence
Tissue sections of whole liver isolated from CaMKK2f/f (n = 3) and CaMKK2LKO (n = 6) mice from Figure 1A were deparaffinized and rehydrated using the standard protocol of immersion in xylene (2 × 5 min, transfer to 100% EtOH (2 × 1 min), transfer to 95% EtOH (1 min), transfer to 70% EtOH (1 min), transfer to 50% EtOH (1 min), transfer to dH2O (2 × 1 min), blot excess water and draw a hydrophobic barrier. A solution of 3% H2O2 in methanol was applied to the sections for 15 min at RT, followed by washing with dH2O (2 × 5 min). Sections were permeabilized with 1% normal donkey serum in PBS-T (2 × 10 min) and blocked with 5% normal donkey serum in PBS-T for 1 h at RT. A solution of 5% donkey serum in PBS-T was prepared by addition of 500 μl of normal donkey serum and 40 μl of Triton X-100 to 9.46 ml of PBS (no Ca2+ or Mg2+). A solution of 1% donkey serum in PBS-T was prepared by addition of 100 μl of normal donkey serum and 40 μl of Triton X-100 to 9.86 ml of PBS (no Ca2+ or Mg2+).
Primary antibodies (Pex3, rabbit, Novus NBP1-52960; Pex19, mouse, Invitrogen MA5-17266) were diluted at 1:250 in 1% normal donkey serum PBS-T, applied to sections, and incubated O/N at 4°C. The following morning, secondary antibodies, AlexaFluor 594 goat α-rabbit (Pex3) and AlexaFluor488 goat α−mouse (Pex19) were diluted at 1:1000 in 1% normal donkey serum PBS-T for 1 h in the dark at RT. Sections were washed with 1% normal donkey serum PBS-T (2 × 10 min). Finally, nuclear labeling was carried out by diluting DAPI (Hoechst 33342) at 1:1000 in PBS and incubation in the dark at RT for 10 min. Slides were washed twice with PBS (5 min, 20 min) and imaged. Imaging was carried out on a EVOS™ M5000 Cell Imaging System (Invitrogen). Three images per slide/mouse were taken and representative images were selected. Quantification was carried out using ImageJ.
3.6. qPCR
Total mRNA was isolated from liver with EZ-10 Spin Column Total RNA Minipreps Super Kit (Bio Z), per the manufacturer's instructions. Reverse transcription was carried out using 2 μg of RNA with Vilo® Superscript Master Mix (Thermo Fisher), per the manufacturer's instructions. For gene expression analyses, cDNA samples were diluted 5-fold. Exactly 2 μl of diluted cDNA was combined with 18 μl of TaqMan master mix containing the appropriate probe and primer pair as indicated by Roche Assay Design Center. β-actin was used as the internal control. All qPCR assays were run on a One Step-Plus (Thermo-Fisher). All qPCR primer sequences and probe combinations are available upon request.
3.7. Immunoblot
Immunoblot analyses were performed using total liver extracts. In brief, 20 μg of protein was loaded onto a 4–20% gradient SDS-PAGE page gel, separated at 200V for 45 min, transferred using the iBlot system for 4:30 min to nitrocellulose membranes, and blocked in 5% milk in Tris-buffered saline with 0.1% Tween-20 (TSBT). Primary antibodies (Pex3, rabbit, Novus NBP1-52960; Pex19, mouse, Invitrogen MA5-17266; β-actin, HRP conjugated, Cell Signaling 5125S, and CaMKK2, mouse, BD Biosciences 610545) were diluted in 1% milk in TBST at 1:1000, except for CaMKK2, which was used at a dilution of 1:400. Nitrocellulose membranes were incubated with primary antibodies overnight with rotation at 4°C. Immunoblots were incubated with appropriate secondary antibodies coupled to horseradish peroxidase (Digital anti-Rabbit-HRP, R1006, 1:1000, Kindle Biosciences and Digital anti-Mouse-HRP, R1005, 1:1000, Kindle Biosciences), reacted with ECL reagents per the manufacturer's instructions (Thermo), and detected by autoradiography or KwikQuant® Imager (Kindle Biosciences).
3.8. In vitro kinase assay
HDMTC buffer was prepared by combining HEPES (25 mM), DDT (0.5 mM), Tween-20 (0.1%), MgCl2 (10 mM), and CaCl2 (1 mM) in nuclease free water. An appropriate amount of HDMTC (45 μl/reaction) was used to make HDMTCC by adding calmodulin (bovine, 1 μM). Another aliquot of HDMTC was reserved for generation of the ATP solution (2 μl/reaction), which was composed of ATP (50 μM) and P32-γ-ATP (0.05 μCi/ml). CaMKK2 (0.5 μg), GAPDH (1 μg), AMPK1-312 (1 μg), CaMKI (1 μg), Pex3 (1 μg), and/or PGK (1 μg), and GST (1 μg) were added to 1.5 ml Eppendorf tubes according to the specific reaction. Exactly 45 μl of HDMTCC was added to each tube and nuclease free water was added to bring the total volume to 50 μl. Tubes were mixed and centrifuged briefly before spiking in the ATP and incubating at 30 °C for 60 min, unless otherwise indicated. For kinetic time course experiments, 10 μl of each reaction was removed from the appropriate tube and added to a tube containing 4 μl of 4X Lamelli buffer to stop the reaction. Samples were denatured at 95 °C for 6 min prior to separation by SDS-PAGE.
Samples were centrifuged and 10 μl were loaded on a 4–20% gradient SDS-PAGE gel and run at 200V until the dye front was no longer visible (approximately 45 min). The gel was removed and fixed in rapid fixing solution (25% 2-propanol, 10% acetic Acid) at RT for 1 h on a rocker. The gel was transferred to rapid Coomassie stain solution (10% acetic Acid, 0.006% Coomassie R250) for 1 h at room temperature on a rocker to visualize bands and transferred to destaining solution (10% Acetic Acid) at room temperature overnight on a rocker to remove background staining. The gel was transferred to Gel Drying Solution (10% Acetic Acid, 30% Methanol, 5% Glycerol) for 1.5 h on a rocker at RT. The gel was then placed on a piece of filter paper, wrapped in plastic wrap, and dried at 50°C for 2 h. The dried gel was placed in a developing cassette, along with film, and placed in the −80°C freezer for 24 h.
3.9. GTT, ITT, PTT
For glucose tolerance tests (GTT) with insulin measurement, mice were fasted for 16 h and administered a 50% glucose solution via IP injection at 2.0 g/kg. Blood samples were collected prior to and after glucose injection at times 0, 15, 30, 60, and 120 min, respectively via tail vein bleeding using a lancet. Blood glucose was measured based on the glucose hexokinase assay and plasma levels of insulin were determined by ELISA (Millipore). Mice were restrained repeatedly for less than a minute each time blood samples were collected.
For insulin tolerance tests (ITT), mice were fasted for 4 h and administered a defined dose of insulin, 1.0 U/kg body weight (Humulin R), via IP injection. Blood glucose was measured at 0 (before) and after 15, 30, 60 and 120 min of insulin injection using a glucometer (Life Scan).
For pyruvate tolerance tests (PTT), mice were fasted for 16 h and administered a defined dose of sodium pyruvate, 2.0 g/kg body weight, via IP injection. Blood glucose was measured at 0 (before) and after 15, 30, 60 and 120 min of insulin injection using a glucometer (Life Scan).
3.10. Combinatorial peptide library screen
The peptide library contained 182 peptide mixtures with the general sequence Y-A-x-x-x-x-x-S/T-x-x-x-x-A-G-K-K(biotin), where x indicates an equimolar mixture of 17 amino acids excluding Cys, Ser and Thr and S/T is a 1:1 mixture of Ser and Thr. Each individual mixture included a single one of the 20 amino acids fixed at one of the x positions. Peptides (50 μM) were arrayed in 1,536-well plates in 2 μl reactions containing 50 mM HEPES, pH 7.4, 10 mM MgCl2, 1 mM CaCl2, 2.5 mM DTT, 0.1% Tween 20, 10 μg/ml human CaMKK2 catalytic domain, 10 μg/ml calmodulin, and 50 μM [γ-32P]-ATP (0.05 μCi/μl). Plates were incubated 2 h at 30 °C, and 200 nl aliquots were transferred to streptavidin membrane (Promega), which was washed, dried, and exposed to a phosphor imaging screen as described [52]. The screen was analyzed by phosphoimager, spot intensities were quantified (BioRad QuantityOne software), and data normalized to the average signal at a given position. Heat maps (Microsoft Excel) show averaged log2 transformed data from two independent experiments.
3.11. Plasma profiling
Whole blood was collected and transferred into a prechilled microcentrifuge tube containing 10 μl of 0.5 M EDTA (pH 8.0) for isolation of EDTA-plasma, which was transferred to a clean microcentrifuge tube and stored at −80 °C until used. Mouse plasma TG, glycerol, FFA, adiponectin, and liver function (i.e., AST/ALT) parameters were measured by the mouse Metabolism Core Services at Baylor College of Medicine.
3.12. Measurement of plasma glucose
Plasma glucose level was measured by a handheld glucometer.
3.13. Liver profiling
Approximately 20 mg pieces of liver were used to measure l-Carnitine (Biovision, #K412-100), long chain free fatty acids (Biovision, #K612-100), PGK activity (AbCam, ab252890) and GAPDH activity (AbCam, ab204732) according to the manufacturer's protocol. Data were normalized to total protein concentration.
3.14. Lipidomics
The extraction of lipids from liver samples and analysis by mass spectrometry were described previously [[53], [54], [55]]. Data acquisition was performed in both positive and negative ionization modes using a TripleTOF 5600 equipped with a Turbo VTM ion source (AB Sciex, Concord, Canada), and earlier detailed methods were described [[53], [54], [55]]. Relative abundance was quantified as the log2 transform of peak areas, and each compound normalized using a spiked internal standard. Quality control samples were used to monitor the overall integrity of the lipid extraction and mass spectrometry analyses. The distributions of detected lipid species across the quality control samples indicated high reproducibility.
3.15. RNA-seq (gene ontology, GSEA, Kaplan–Meier plots)
RNA samples were isolated from CaMKK2f/f, CaMKK2LKO, CaMKK2−/− + AAV9-GFP, and CaMKK2−/− + AAV9-CaMKK2 liver tissues using the miRNeasy Mini Kit (Qiagen) per the manufacturer's instructions. RNA samples (at least 2000 ng of total RNA for each sample) were processed by NovoGene® for library prep, quality assurance, and sequenced with an average of 20 million paired end reads/sample.
Gene Set Enrichment Analysis (GSEA) was carried out using the GSEA software package [56] to assess the degree of similarity among the studied gene signatures [57,58]. For the CaMKK2LKO transcriptome response, all genes were ranked by the fold-change between the CaMKK2LKO and the CaMKK2f/f sample groups. Normalized Enrichment Score (NES) and adjusted q-values were computed utilizing the GSEA method, based on 1000 random permutations of the ranked genes as previously described [32,59,60]. Enriched pathways were determined using the gene set collections at the Molecular Signature Database (MSigDB, http://www.broadinstitute.org/gsea/msigdb/) using a hypergeometric distribution (p < 0.05).
Survival analysis correlating CaMKK2LKO gene signatures was performed by sorting all the patient samples [61,62] based on the individual expression or signature scores. The survival or disease-free survival Kaplan–Meier plots were generated using “survival” package in the R statistical system. Significance was determined by log-rank test (weights 0 or 4, respectively).
3.16. CLAMs (indirect calorimetry)
Calorimetry (Columbus Instruments) was used for real-time measurement of Respiratory Exchange Ratio (RER). Mice fed ad libitum were acclimated to the chambers for at least one week and food intake was monitored for 5 days under a 12-hour light/dark cycle. CalR (version 1.2, https://calrapp.org) [63] was used for the analysis of experiments using indirect calorimetry to measure physiological energy balance. Data were collected for VO2 consumed and VCO2 released, and RER was calculated as VCO2/VO2.
3.17. Seahorse extracellular flux analysis
Primary hepatocytes isolated from CaMKK2f/f and CaMKK2LKO whole liver were plated on XF96 well cell culture microplates (Agilent) pre-coated with 1:60 dilution of a 3 mg/ml rat tail collagen:HBSS solution. Hepatocytes were plated at a density of 80,000 cells/well to form a consistent and confluent monolayer at the time of experimental measurements. Extracellular flux analysis was performed using the Seahorse XF cell mitochondrial stress test kit (Seahorse Bioscience P/N 103015-100). Experimental medium was XF DMEM supplemented with glucose (10 mM), pyruvate (1 mM) and glutamine (2 mM), unless noted otherwise. The final concentration of oligomycin used in the experiment was 2 μM, FCCP was 2 μM and rotenone/antimycin was 0.5 μM. Extracellular flux experiments were performed on a Seahorse XF96 Analyzer (Agilent).
3.18. β-oxidation assay
β-oxidation assays were performed according to Akie & Cooper [64]. In short, 12-well plates were coated the day prior to primary hepatocyte isolation by adding 300 μl of a 1:60 dilution of a 3 mg/ml rat tail collagen:HBSS solution and incubated at room temperature for 2 h. Collagen was removed by aspiration and plates air dried overnight. Plates were washed with HBSS prior to plating of primary hepatocytes. Primary hepatocytes were plated in William's E media at a density of 400,000 cells/well and allowed to adhere for 6 h. Media was then changed to serum starvation medium (DMEM no glucose, 0.2% BSA Fraction V, 2 mM sodium pyruvate, 2% Pen/Strep, and 20 nM glucagon) and cells were incubated at 37°C for 16 h. Serum Starvation Medium was removed and 1 ml of Pre-Incubation Medium (DMEM no glucose, 25 mM HEPES, 1% BSA Fraction V, 250 mM of sodium palmitate, and 20 nM glucagon) was added and incubated at 37°C for 2 h. DMSO, STO-609 (20 mM), etomoxir (20 mM), enoximone (150 mM), or oligomycin (10 mM) were added to the appropriate wells at this time.
During the incubation period, the appropriate amount of 0.1 μCi/ml 14C-palmitate (0.5 μCi/well) was dried under air. Dried palmitate was resuspended in 0.1 N NaOH to a final concentration of 12.5 ml/μCi and incubated at 70°C for 10 min followed by the addition of 3 volumes of Pre-Incubation Media to the palmitate and mixed via pipetting. At the end of the incubation period, 25 μl of diluted 14C-palmitate was spiked into each well and cells incubated for 90 min. During this incubation, filter paper was added to the bottom of a new 12-well plate such that it completely covered the bottom surface of each well. Parafilm was used to cover the plate and a pipette tip was used to cut out the well openings. Next, 10 min before the incubation period ended, 100 μl of 3N NaOH was added to each well of the filter paper plate, ensuring complete absorption of liquid. The cell plate was snap frozen in liquid nitrogen and each well was checked for complete freezing. Next, 200 μl of 70% perchloric acid was added to each well and the filter plate was immediately placed on top. Plates were wrapped together with parafilm and rocked at room temperature for 16 h. The following day, the filter paper was removed and added to 4 ml of liquid scintillation fluid in a scintillation vial and 14C counts were measured on a Beckmann LS6500 scintillation counter.
3.19. GST-CaMKK2 pull-down
A cold in vitro kinase reaction was performed as previously described. All reactions contained GST-CaMKK2, except for the GST control reaction. Exactly 20 μl of the kinase reaction was added to a new 1.5 ml Eppendorf tube. To immobilize GST-CaMKK2 or GST alone, 20 μl of a 50/50 slurry of glutathione-sepharose beads (50% bead, 50% IP affinity precipitation buffer) was added to each tube and brought to a final volume of 500 μl using IP affinity precipitation buffer (50 mM Tris–HCl 7.5, 150 mM NaCl, 10% glycerol, and 1% IGEPAL), followed by overnight rotation at 4°C. Reactions were then centrifuged at 1,000 rcf for 30 s to pellet the beads and the supernate was removed and discarded. Beads were washed three times with 500 μl of affinity precipitation buffer per wash, followed by centrifugation at 1,000 rcf for 30 s. Exactly 40 μl of loading buffer was added to the beads after the final wash and denatured at 95°C for 6 min. A total of 20 μl from each reaction was separated by SDS-PAGE and immunoblotting was performed as previously described.
3.20. Immunoprecipitation of Pex3
A cold in vitro kinase was performed as previously described. Interaction of Pex3 and Pex19 was interrogated in the presence and absence of CaMKK2 in various activation states. Exactly 20 μl of the total kinase reaction was added to a new 1.5 ml Eppendorf tube. The total volume was brought to 1 ml with IP affinity precipitation buffer. Next, 2 μg of Pex3 antibody was added, except for the bead only control. Reactions were incubated at 4°C with rotation for 2 h, after which 25 μl of Protein A/G PLUS-Agarose (Santa Cruz, sc-2003) was added followed by overnight rotation at 4°C. Reactions were then centrifuged at 1,000 rcf for 30 s to pellet the beads and the supernate was removed and discarded. Beads were washed three times with 500 μl of IP affinity precipitation buffer per wash, followed by centrifugation at 1,000 rcf for 30 s. Exactly 40 μl of loading buffer was added to the beads after the final wash and denatured at 95°C for 6 min. A total of 20 μl from each reaction was separated by SDS-PAGE and immunoblotting was performed as previously described. The following antibodies were used for IP: Pex3 (Novus, NBP1-52960; Pex19, mouse, Invitrogen MA5-17266).
3.21. Peroxisomal isolation and purification
Isolation of functional peroxisomes was carried out using a published protocol from Sigma (Perox1) [65] with minor modification as previously described [45]. Briefly, CaMKK2f/f (n = 4) and 4 CaMKK2LKO (n = 4) male mice were euthanized with isofluorane and livers were harvested such that 4 g of liver tissue were collected from each genotype (1 g/mouse) and combined for isolation of peroxisomes. All reagents were ice cold unless otherwise noted, and all steps were performed on ice. The crude peroxisome fraction (CPF) was obtained by cutting the liver tissue using a razor blade in a petri dish into small pieces. Liver pieces were transferred to a prechilled glass Dounce® homogenizer and homogenized with 15 strokes of the glass pestle. The Sigma protocol was followed as outlined until the last centrifugation step, which was carried out at 14,000×g for 35 min due to speed restrictions of the rotor. The conversion was carried out using the formula:
where is the new spin time, is the protocol spin time, is the protocol spin speed, and is the new spin speed. Purification of the CPF was carried out according to the Sigma protocol. Per the protocol, the bottom layer was collected (∼1 ml), which represents the purified peroxisomal fraction (PPF). Due to the presence of mitochondrial contamination, we utilized the magnetic-activated cell sorting (MACS) column purification system. The PPF was incubated with 2 μl of biotinylated carnitine palmitoyl transferase 1B (Cpt1B, BiossUSA.com, bs-5045R-biotin) antibody for 10 min at a 1:500 dilution. Next, anti-biotin microbeads (MiltenyiBiotec 130-090-485) were added to the PPF. The resulting suspension was applied to prepped LD columns (MiltenyiBiotec 130-042-901) and the flowthrough was collected and saved.
3.22. Peroxisomal Seahorse assay
The peroxisomal Seahorse assay was carried out as previously described [45]. Briefly, Seahorse Base Assay Media was supplemented with 100 μM ATP. A total of 3% BSA was added to the control media and 3% of long chain fatty acids (i.e., palmitate, oleate, and linolate) were added to the experimental media at a concentration of 1% each.
The calibration plate was set up the night before according to the Seahorse protocol. The run included 6 measurement cycles, 4 min each, with no mixing or injections. Four blank wells were programmed as internal negative controls. During the calibration step, 50 μg of peroxisomes was plated on ice in the Seahorse cell plate and then centrifuged at 12,000×g for 15 min at 4 °C. The appropriate media was added to each well to a final volume of 125 μl and the plate kept on ice until the start of the assay.
3.23. In-vitro GAPDH activity assay
A cold kinase assay was performed as previously described, except 5 ng of GAPDH was added to the indicated reaction tubes and reactions were allowed to incubate for 1.5 h. Following incubation, the total reaction volume was added to the GAPDH activity assay (AbCam, ab204732) according to the manufacturer's protocol.
3.24. Stable isotope tracer experiments
Primary hepatocytes were isolated from CaMKK2f/f and CaMKK2LKO mice and plated in William's E media on Primaria 15-cm tissue culture dishes (Corning) at a density of 5 million/plate and allowed to adhere at 37°C for 6 h. Media was changed to starvation media (DMEM, no glucose) and cells were incubated for 16 h at 37°C. Dialyzed BSA (10%) and Pen/Strep (1%) were added to all plates. For the 13C-glucose experiments, 12 mM of 13C-glucose (Cambridge Isotope Laboratories CLM-1396, n = 4/genotype) or 12C-glucose (Thermo A2494001, n = 1/genotype) was added to the cells and incubated at 37°C for 6 h (short) or 72 h (long). For the 13C-palmitate experiments, 1% BSA Fraction V was added to all plates, along with either 150 μM of 13C-palmitate (CLM3943, n = 4/genotype) or 150 μM of 12C-palmitate (Sigma P9767, n = 4/genotype). Metabolic extraction from cell pellets was performed as described previously [66]. Fatty acid 13C incorporation was measured using a Luna 3 μm Phenyl-Hexyl column (150 × 2 mm; (Phenomenex, Torrance, CA). The mobile phases A and B were 10 mM ammonium acetate (pH 8) and methanol. Gradient Flow: 0–8 min 40% B, 8–13 min 50% B, 13–23 min 67%, 23–30 min 100%, and 30 min 40%, followed by re-equilibration until the end of the gradient, approximately 37 min prior to the initial starting condition of 40% B. Flow rate of the solvents used for analysis was 0.2 ml/min. The injection volume was 20 μL. The above mentioned sample volumes were injected and acquired the data via multiple reaction monitoring (MRM) in negative mode using a 6495 Triple Quadrupole mass spectrometry coupled to an Agilent UHPLC system (Agilent Technologies, Santa Clara, CA) through Agilent Mass Hunter Software [67]. The acquired data were analyzed, and integration of each peak was performed using Agilent Mass Hunter Quantitative Analysis software. Percentage of fatty acid 13C incorporation was calculated using Microsoft excel from peak area and represented as bar graphs.
3.25. Statistical analyses
Statistical testing was performed using a two-tailed Student's t-test unless indicated otherwise. Area under the curve and repeated measures ANOVA were performed to compare two genotype groups analyzed with time (GTT, ITT, PTT, Seahorse). Data were tested for normality and equal variance assumptions and log transformed prior to analysis when necessary. This approach reduced the total number of mice required per IACUC standards and all experiments adhered to ARRIVE guidelines.
4. Discussion
The current study was designed to evaluate the mechanistic role of hepatic CaMKK2 in the development of peripheral insulin resistance and to determine its function in the regulation of hepatic metabolism. Consistent with previously published work [15,16], our findings demonstrate that liver-specific ablation of CaMKK2 confers protection against insulin resistance when challenged with HFD and this effect is reversed upon re-expression of hepatic CaMKK2. Earlier studies from our laboratories demonstrated that either genetic or acute loss of CaMKK2 improves insulin sensitivity when mice are challenged with HFD [15,16], but failed to provide a definitive mechanism to explain this phenotype. Furthermore, our previous work established CaMKK2 as a potential fuel source selector given the robust phenotypic switch in hepatic steatosis observed in whole-body CaMKK2 knockouts when challenged with either HFD or HCD diets [16]. Mirroring these phenotypic observations, we found that hepatocytes devoid of CaMKK2 are unable to metabolize glucose but show improvements in fatty acid oxidation compared to wildtype. We identified GAPDH and PEX3 as two novel phospho-targets of CaMKK2 that contribute to the mechanisms responsible for this metabolic shift. We propose that CaMKK2 promotes peripheral insulin resistance by metabolically reprogramming the liver to favor glucose over lipid metabolism, an alteration that has gained recent attention for its contributions to the development of systemic metabolic disease [14,68,69]
CaMKK2 is a well-established molecular activator and regulator of AMP-dependent protein kinase (AMPK) [70]. CaMKK2 directly phosphorylates AMPK at T172, a mark that has been implicated in inflammation, neuronal ischemic injury, and satiety [[71], [72], [73]]. Although CaMKK2 activity is certainly a determinant of AMPK action, the overarching metabolic phenotypes of our CaMKK2LKO mice fail to phenocopy the hepatic loss of AMPK [74,75]. In contrast with our findings in CaMKK2LKO mice fed HFD, AMPKa1/a2LKO mice challenged with HFD failed to show significant differences in steady state blood glucose, response to glucose challenge (GTT), plasma insulin and dyslipidemia. While these observations suggest that hepatic CaMKK2 function is not entirely contingent upon AMPK activity, our findings highlight a more elegant mechanism by which CaMKK2 may intersect with the pleiotropic metabolic functions of AMPK. In short, the role of AMPK in the context of metabolic homeostasis is to actively interpret the balance of ATP catabolism by regulating programs that replenish the energy-depleted state while simultaneously attenuating energy-consuming processes [76,77]. Our discovery that CaMKK2 functions to balance glycolytic and fatty acid metabolism for ATP production exposes an overarching regulatory mechanism that links the molecular activation of CaMKK2 to provide the obligate energetic requirements that feed downstream AMPK-dependent pathways to maintain metabolic homeostasis. Such a molecular hierarchy ensures direct coordination and integration between energy producing pathways controlled by CaMKK2 that sponsor and fuel the metabolic actions of AMPK.
Given that none of the established targets of CaMKK2 fully explain its complex phenotypic and cellular metabolic functions, we set out to identify additional molecular effectors that better clarify its roles for balancing glycolytic and fatty acid metabolism. We found that CaMKK2 directly phosphorylates and stimulates GAPDH activity to promote glycolytic metabolism (Figure 3F–H). This newly defined mechanism provides a plausible explanation for why CaMKK2−/− mice develop hepatic steatosis when challenged with a high carbohydrate diet [16]. In the context of fatty acid metabolism, CaMKK2LKO hepatocytes metabolize lipid more efficiently through engagement of peroxisomal β-oxidation. We found that CaMKK2 directly interacts with and phosphorylates the peroxisomal biogenesis factor, Pex3 (Figure 4K). Pex3 functions in partnership with its cytosolic partner Pex19 to form new peroxisomal membranes as well as lipid droplets [29,49]. Interestingly, Pex3 association with Pex19 regulates the trafficking of dual-targeted proteins that reside in the membranes of peroxisomes and mitochondria [49]. Taken together, our discovery of a new CaMKK2-Pex3 signaling axis establishes a molecular pathway for the regulation of peroxisomal lipid metabolism for pre-processing long and very-long chain fatty acids to more readily available forms that bypass the mitochondrial CPT1 transport system. We contend this mechanism is partially responsible for the increased fatty acid oxidation observed in CaMKK2LKO hepatocytes (Figure 4A) and the improved metabolic phenotypes of CaMKK2LKO mice challenged with HFD (Figure 1).
In summary, our current study exposes two alternative mechanisms by which CaMKK2 influences the balance of hepatic metabolism: 1) via GAPDH to promote glycolysis and 2) via PEX3 to suppress fatty acid oxidation. While our study identified a functional interplay between CaMKK2 and these putative substrates, additional research is needed to validate these proteins as endogenous phosphotargets and to identify the CaMKK2 specific phosphorylation sites on each. Our findings demonstrate that suppression of CaMKK2 activity in the liver blunts glucose metabolism and bolsters lipid catabolism, resulting in significant improvements to peripheral insulin sensitivity and hepatic health under conditions of HFD feeding. While genetic ablation or pharmacological inhibition of hepatic CaMKK2 appears to support improved peripheral metabolic health when fed excess fat, dietary overload with carbohydrate has the opposite effect. This paradoxical phenotype in CaMKK2LKO mice is accompanied by an unexplained incorporation of glycolytic carbon into long chain fatty acids. These findings highlight the intricate regulatory connections for how lipid and glycolytic carbon are metabolized, utilized and even exchanged and warrant further study to uncover new CaMKK2-dependent conduits and connections between these pathways. Collectively, we have linked the hepatic functions of CaMKK2 to molecular mechanisms that regulate metabolic homeostasis through organelle crosstalk to control the fate of glycolytic and fatty acid carbon. These results suggest that pharmacological regulation of CaMKK2 may enable metabolic reprogramming of the liver to favor peroxisomal β-oxidation of fat as a potential therapeutic treatment for metabolic liver disease and peripheral insulin resistance.
Acknowledgements
We acknowledge the joint participation by the Adrienne Helis Malvin Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with Baylor College of Medicine and the CaMKK2-PKM2 promotes HCC research program. We thank and acknowledge the support of the Mouse Metabolism and Phenotyping Core at Baylor College of Medicine (NIH-R01DK114356 & UM1HG006348), the Gene Vector Core at Baylor College of Medicine, the Metabolomics Core at Baylor College of Medicine (CPRIT Core Facility Support Award RP170005, NIH-P30 CA125123), the Metabolomics Core Facility at MD Anderson (NIH S10OD012304-01 and P30CA016672), the Dan L. Duncan Cancer Center, the Pathology and Histology Core at Baylor College of Medicine (NCI-CA125123), and the Biostatistics and Informatics Shared Resource Core at Baylor College of Medicine. We would also like to personally thank Peter Masschelin, M.S. at Baylor College of Medicine for his assistance with CalR calculations. Additionally, we thank Naomi Gonzales, Vasanta Putluri, Chandra Ambati, and Sandra Grimm for their technical expertise and contributions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101513.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Figs1.
Figs2.
Figs3.
Figs4.
References
- 1.Fu S., Yang L., Li P., Hofmann O., Dicker L., Hide W., et al. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fabbrini E., Sullivan S., Klein S. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology. 2010;51:679–689. doi: 10.1002/hep.23280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serfaty L., Lemoine M. Definition and natural history of metabolic steatosis: clinical aspects of NAFLD, NASH and cirrhosis. Diabetes & Metabolism. 2008;34:634–637. doi: 10.1016/S1262-3636(08)74597-X. [DOI] [PubMed] [Google Scholar]
- 4.Duwaerts C.C., Maher J.J. Macronutrients and the adipose-liver Axis in obesity and fatty liver. CMGH. 2019:7 749–761. doi: 10.1016/j.jcmgh.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mu W., Xuefang C., Liu Y., Qianzhou L., Gaolin L., Jigang Z., et al. Potential nexus of non-alcoholic fatty liver disease and type 2 diabetes mellitus: insulin resistance between hepatic and peripheral tissues. Frontiers in Pharmacology. 2019;9:1566. doi: 10.3389/fphar.2018.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayiner M., Koenig A., Henry L., Younossi Z.M. Epidemiology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis in the United States and the rest of the world. Clinics in Liver Disease. 2016;20:205–214. doi: 10.1016/j.cld.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Ertle J., Dechêne A., Sowa J.-P., Penndorf V., Herzer K., Kaiser G., et al. Non-alcoholic fatty liver disease progresses to hepatocellular carcinoma in the absence of apparent cirrhosis. International Journal of Cancer. 2011;128:2436–2443. doi: 10.1002/ijc.25797. [DOI] [PubMed] [Google Scholar]
- 8.Malik S.M., deVera M.E., Fontes P., Shaikh O., Ahmad J. Outcome after liver transplantation for NASH cirrhosis. American Journal of Transplantation. 2009;9:782–793. doi: 10.1111/j.1600-6143.2009.02590.x. [DOI] [PubMed] [Google Scholar]
- 9.Wong R.J., Aguilar M., Cheung R., Perumpail R.B., Harrison S.A., Younossi Z.M., et al. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology. 2015;148:547–555. doi: 10.1053/j.gastro.2014.11.039. [DOI] [PubMed] [Google Scholar]
- 10.Konerman M.A., Jones J.C., Harrison S.A. 2017. Pharmacotherapy for NASH: current and emerging. [DOI] [PubMed] [Google Scholar]
- 11.Rui L. Energy metabolism in the liver. Comprehensive Physiology. 2014;4:177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trefts E., Gannon M., Wasserman D.H. The liver. Current Biology. 2017;27:R1147–R1151. doi: 10.1016/j.cub.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klover P.J., Mooney R.A. Hepatocytes: critical for glucose homeostasis. The International Journal of Biochemistry & Cell Biology. 2004;36:753–758. doi: 10.1016/j.biocel.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Rutkowski D.T. Liver function and dysfunction - a unique window into the physiological reach of ER stress and the unfolded protein response. FEBS Journal. 2018 doi: 10.1111/febs.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson K.A., Lin F., Ribar T.J., Stevens R.D., Muehlbauer M.J., Newgard C.B., et al. 2012. Deletion of CaMKK2 from the liver lowers blood glucose and improves whole-body glucose tolerance in the mouse. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcelo K.L., Ribar T., Means C.R., Tsimelzon A., Stevens R.D., Ilkayeva O., et al. Research Resource: roles for calcium/calmodulin-dependent protein kinase kinase 2 (CaMKK2) in systems metabolism. Molecular Endocrinology. 2016;30:557–572. doi: 10.1210/me.2016-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.York B., Li F., Lin F., Marcelo K.L., Mao J., Dean A., et al. Pharmacological inhibition of CaMKK2 with the selective antagonist STO-609 regresses NAFLD. Scientific Reports. 2017;7:11793. doi: 10.1038/s41598-017-12139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houten S.M., Wanders R.J.A., Ranea-Robles P. Metabolic interactions between peroxisomes and mitochondria with a special focus on acylcarnitine metabolism. Biochimica et Biophysica Acta - Molecular Basis of Disease. 2020;1866:165720. doi: 10.1016/j.bbadis.2020.165720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flis V.V., Daum G. Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harbor Perspectives in Biology. 2013;5 doi: 10.1101/cshperspect.a013235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartlett K., Eaton S. Mitochondrial β-oxidation. European Journal of Biochemistry. 2004;271:462–469. doi: 10.1046/j.1432-1033.2003.03947.x. [DOI] [PubMed] [Google Scholar]
- 21.Islinger M., Voelkl A., Fahimi H.D., Schrader M. The peroxisome: an update on mysteries 2.0. Histochemistry and Cell Biology. 2018;150:443–471. doi: 10.1007/s00418-018-1722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lodhi I.J., Semenkovich C.F. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metabolism. 2014;19:380–392. doi: 10.1016/j.cmet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noland R.C., Koves T.R., Seiler S.E., Lum H., Lust R.M., Ilkayeva O., et al. Carnitine insufficiency caused by aging and overnutrition compromises mitochondrial performance and metabolic control. Journal of Biological Chemistry. 2009;284:22840–22852. doi: 10.1074/jbc.M109.032888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wicks S.E., Vandanmagsar B., Haynie K.R., Fuller S.E., Warfel J.D., Stephens J.M., et al. Impaired mitochondrial fat oxidation induces adaptive remodeling of muscle metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E3300–E3309. doi: 10.1073/pnas.1418560112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashimoto T., Fujita T., Usuda N., Cook W., Qi C., Peters J.M., et al. Peroxisomal and mitochondrial fatty acid β-oxidation in mice nullizygous for both peroxisome proliferator-activated receptor and peroxisomal fatty acyl-CoA oxidase: genotype correlation with fatty liver phenotype. Journal of Biological Chemistry. 1999;274:19228–19236. doi: 10.1074/jbc.274.27.19228. [DOI] [PubMed] [Google Scholar]
- 26.Wicks S.E., Vandanmagsar B., Haynie K.R., Fuller S.E., Warfel J.D., Stephens J.M., et al. Impaired mitochondrial fat oxidation induces adaptive remodeling of muscle metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E3300–E3309. doi: 10.1073/pnas.1418560112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Violante S., Achetib N., van Roermund C.W.T., Hagen J., Dodatko T., Vaz F.M., et al. Peroxisomes can oxidize medium- and long-chain fatty acids through a pathway involving ABCD3 and HSD17B4. Federation of American Societies for Experimental Biology Journal. 2019;33:4355–4364. doi: 10.1096/fj.201801498R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang C.L., Weigel A.V., Ioannou M.S., Amalia Pasolli H., Shan Xu C., Peale D.R., et al. Spastin tethers lipid droplets to peroxisomes and directs fatty acid trafficking through ESCRT-III. Journal of Cell Biology. 2019;218:2583–2599. doi: 10.1083/jcb.201902061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joshi A.S., Cohen S. Lipid droplet and peroxisome biogenesis: do they go hand-in-hand? Frontiers in Cell and Developmental Biology. 2019;7:1–9. doi: 10.3389/fcell.2019.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson, K.A., Ribar, T.J., Lin, F., Noeldner, P.K., Green, M.F., Muehlbauer, M.J. et al. Hypothalamic CaMKK2 contributes to the regulation of energy balance. doi:10.1016/j.cmet.2008.02.011. [DOI] [PubMed]
- 31.Chen B.D., He C.H., Chen X.C., Pan S., Liu F., Ma X., et al. Targeting transgene to the heart and liver with AAV9 by different promoters. Clinical and Experimental Pharmacology and Physiology. 2015;42:1108–1117. doi: 10.1111/1440-1681.12453. [DOI] [PubMed] [Google Scholar]
- 32.Lin F., Marcelo K.L., Rajapakshe K., Coarfa C., Dean A., Wilganowski N., et al. The CaMKK2/CaMKIV relay is an essential regulator of hepatic cancer. Hepatology. 2015;62:505–520. doi: 10.1002/hep.27832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berg J.M., Tymoczko J.L., Stryer L. 2002. Glycolysis and gluconeogenesis. [Google Scholar]
- 34.Ma D.W.L., Arendt B.M., Hillyer L.M., Fung S.K., McGilvray I., Guindi M., et al. Plasma phospholipids and fatty acid composition differ between liver biopsy-proven nonalcoholic fatty liver disease and healthy subjects. Nutrition & Diabetes. 2016;6 doi: 10.1038/nutd.2016.27. 2016 6:7. e220–e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z., A L., A. T., U M., J L., M A. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metabolism. 2006;3:321–331. doi: 10.1016/j.cmet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Arendt B.M., M D., S B., N S., T G., G M. Non-alcoholic fatty liver disease is associated with lower hepatic and erythrocyte ratios of phosphatidylcholine to phosphatidylethanolamine. Applied Physiology Nutrition and Metabolism. 2012;38:334–340. doi: 10.1139/apnm-2012-0261. [DOI] [PubMed] [Google Scholar]
- 37.Bonekamp N.A., Völkl A., Fahimi H.D., Schrader M. Reactive oxygen species and peroxisomes: struggling for balance. BioFactors. 2009;35:346–355. doi: 10.1002/biof.48. [DOI] [PubMed] [Google Scholar]
- 38.Hunt M.C., Siponen M.I., Alexson S.E.H. The emerging role of acyl-CoA thioesterases and acyltransferases in regulating peroxisomal lipid metabolism. Biochimica et Biophysica Acta - Molecular Basis of Disease. 2012;1822:1397–1410. doi: 10.1016/j.bbadis.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Wanders R.J.A. Metabolic functions of peroxisomes in health and disease. Biochimie. 2014;98:36–44. doi: 10.1016/j.biochi.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 40.Reddy J.K., Hashimoto T. P eroxisomal β-O xidation and P eroxisome P roliferator –a ctivated R eceptor α: an adaptive metabolic system. Annual Review of Nutrition. 2002;21:193–230. doi: 10.1146/annurev.nutr.21.1.193. [DOI] [PubMed] [Google Scholar]
- 41.Hashimoto T. Individual peroxisomal βT-OXIDATION enzymes. Annals of the New York Academy of Sciences. 1982;386:5–12. doi: 10.1111/j.1749-6632.1982.tb21403.x. [DOI] [PubMed] [Google Scholar]
- 42.Schrader M., Kamoshita M., Islinger M. Organelle interplay—peroxisome interactions in health and disease. Journal of Inherited Metabolic Disease. 2020;43:71–89. doi: 10.1002/jimd.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wanders R.J.A., Komen J., Kemp S. Fatty acid omega-oxidation as a rescue pathway for fatty acid oxidation disorders in humans. FEBS Journal. 2011;278:182–194. doi: 10.1111/j.1742-4658.2010.07947.x. [DOI] [PubMed] [Google Scholar]
- 44.Abdel-Aleem S., Youssef J., Frangakis C., Badr M. Selective inhibition of hepatic peroxisomal fatty acid beta-oxidation by enoximone. Life Sciences. 1992;51:53–57. doi: 10.1016/0024-3205(92)90218-e. [DOI] [PubMed] [Google Scholar]
- 45.Stork B., Dean A., York B. Methodology for measuring oxidative capacity of isolated peroxisomes in the Seahorse assay. Journal of Biological Methods. 2022;9 doi: 10.14440/jbm.2022.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H., de Boer R., Krikken A.M., Akşit A., Yuan W., van der Klei I.J. Peroxisome development in yeast is associated with the formation of Pex3-dependent peroxisome-vacuole contact sites. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2019;1866:349–359. doi: 10.1016/j.bbamcr.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 47.Colasante C., Chen J., Ahlemeyer B., Bonilla-Martinez R., Karnati S., Baumgart-Vogt E. New insights into the distribution, protein abundance and subcellular localisation of the endogenous peroxisomal biogenesis proteins PEX3 and PEX19 in different organs and cell types of the adult mouse. PLoS One. 2017;12 doi: 10.1371/journal.pone.0183150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erdmann R. Assembly, maintenance and dynamics of peroxisomes. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2016;1863:787–789. doi: 10.1016/j.bbamcr.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 49.Jansen R.L.M., van der Klei I.J. The peroxisome biogenesis factors Pex3 and Pex19: multitasking proteins with disputed functions. FEBS Letters. 2019;593:457–474. doi: 10.1002/1873-3468.13340. [DOI] [PubMed] [Google Scholar]
- 50.McDonnell M.M., Burkhart S.E., Stoddard J.M., Wright Z.J., Strader L.C., Bartel B. The early-acting peroxin PEX19 is redundantly encoded, farnesylated, and essential for viability in arabidopsis thaliana. PLoS One. 2016;11:1–19. doi: 10.1371/journal.pone.0148335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chow F.A., Anderson K.A., Noeldner P.K., Means A.R. The autonomous activity of calcium/calmodulin-dependent protein kinase IV is required for its role in transcription. Journal of Biological Chemistry. 2005 doi: 10.1074/jbc.M500067200. [DOI] [PubMed] [Google Scholar]
- 52.Mock J., Kim P.M., Lam H.Y.K., Piccirillo S., Zhou X., Jeschke G.R., et al. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Science Signaling. 2010;3 doi: 10.1126/scisignal.2000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piyarathna D.W.B., Rajendiran T.M., Putluri V., Vantaku V., Soni T., Rundstedt F.-C.V., et al. Distinct lipidomic landscapes associated with clinical stages of urothelial cancer of the bladder. Eur Urol Focus. 2018;4:907–915. doi: 10.1016/j.euf.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purwaha P., Gu F., Piyarathna D.W.B., Rajendiran T., Ravindran A., Omilian A.R., et al. Unbiased lipidomic profiling of triple-negative breast cancer tissues reveals the association of sphingomyelin levels with patient disease-free survival. Metabolites. 2018;8 doi: 10.3390/metabo8030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vantaku V., Dong J., Ambati C.R., Perera D., Donepudi S.R., Amara C.S., et al. Multi-omics integration analysis robustly predicts high-grade patient survival and identifies CPT1B effect on fatty acid metabolism in bladder cancer. Clinical Cancer Research. 2019;25:3689–3701. doi: 10.1158/1078-0432.CCR-18-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. From the Cover: gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahrens M., Ammerpohl O., Von Schönfels W., Kolarova J., Bens S., Itzel T., et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metabolism. 2013;18:296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 58.Knebel B., Hartwig S., Haas J., Lehr S., Goeddeke S., Susanto F., et al. Peroxisomes compensate hepatic lipid overflow in mice with fatty liver. Biochimica et Biophysica Acta. 2015;1851:965–976. doi: 10.1016/j.bbalip.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 59.Meng H., Gonzales N.M., Lonard D.M., Putluri N., Zhu B., Dacso C.C., et al. XBP1 links the 12-hour clock to NAFLD and regulation of membrane fluidity and lipid homeostasis. Nature Communications. 2020;11:1–16. doi: 10.1038/s41467-020-20028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marcelo K.L., Lin F., Rajapakshe K., Dean A., Gonzales N., Coarfa C., et al. Deciphering hepatocellular responses to metabolic and oncogenic stress. J Biol Methods. 2015;2 doi: 10.14440/jbm.2015.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roessler S., Jia H.L., Budhu A., Forgues M., Ye Q.H., Lee J.S., et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Research. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roessler S., Long E.L., Budhu A., Chen Y., Zhao X., Ji J., et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142 doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mina A.I., LeClair R.A., LeClair K.B., Cohen D.E., Lantier L., Banks A.S. CalR: a web-based analysis tool for indirect calorimetry experiments. Cell Metabolism. 2018;28:656–666. doi: 10.1016/j.cmet.2018.06.019. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Akie T.E., Cooper M.P. Determination of fatty acid oxidation and lipogenesis in mouse primary hepatocytes. Journal of Visualized Experiments. 2015 doi: 10.3791/52982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lazarow P.B., De Duve C. A fatty acyl CoA oxidizing system in rat liver peroxisomes; enhancement by clofibrate, a hypolipidemic drug. Proceedings of the National Academy of Sciences of the United States of America. 1976;73:2043–2046. doi: 10.1073/pnas.73.6.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vantaku V., Putluri V., Bader D.A., Maity S., Ma J., Arnold J.M., et al. Epigenetic loss of AOX1 expression via EZH2 leads to metabolic deregulations and promotes bladder cancer progression. Oncogene. 2020;39:6265–6285. doi: 10.1038/s41388-019-0902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gohlke J.H., Lloyd S.M., Basu S., Putluri V., Vareed S.K., Rasaily U., et al. Methionine-homocysteine pathway in african-American prostate cancer. JNCI Cancer Spectrum. 2019;3 doi: 10.1093/jncics/pkz019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bechmann L.P., Hannivoort R.A., Gerken G., Hotamisligil G.S., Trauner M., Canbay A. The interaction of hepatic lipid and glucose metabolism in liver diseases. Journal of Hepatology. 2012;56:952–964. doi: 10.1016/j.jhep.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 69.Schenk S., Saberi M., Olefsky J.M. Insulin sensitivity: modulation by nutrients and inflammation. The Journal of Clinical Investigation. 2008;118:2992–3002. doi: 10.1172/JCI34260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Green M.F., Anderson K.A., Means A.R. Characterization of the CaMKKβ–AMPK signaling complex. Cellular Signalling. 2011;23:2005–2012. doi: 10.1016/j.cellsig.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y., Xu N., Ding Y., Zhang Y., Li Q., Flores J., et al. Chemerin suppresses neuroinflammation and improves neurological recovery via CaMKK2/AMPK/Nrf2 pathway after germinal matrix hemorrhage in neonatal rats. Brain, Behavior, and Immunity. 2018;70:179–193. doi: 10.1016/j.bbi.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rousset C.I., Leiper F.C., Kichev A., Gressens P., Carling D., Hagberg H., et al. A dual role for AMP-activated protein kinase (AMPK) during neonatal hypoxic–ischaemic brain injury in mice. Journal of Neurochemistry. 2015;133:242–252. doi: 10.1111/jnc.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu W.-N., Wu P.-F., Zhou J., Guan X.-L., Zhang Z., Yang Y.-J., et al. Orexin-A activates hypothalamic AMP-activated protein kinase signaling through a Ca2+-dependent mechanism involving voltage-gated L-type calcium channel. Molecular Pharmacology. 2013;84:876–887. doi: 10.1124/mol.113.086744. [DOI] [PubMed] [Google Scholar]
- 74.Zhao P., Sun X., Chaggan C., Liao Z., Wong K., He F., et al. An AMPK–caspase-6 axis controls liver damage in nonalcoholic steatohepatitis. Science. 2020;367:652–660. doi: 10.1126/science.aay0542. 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boudaba N., Marion A., Huet C., Pierre R., Viollet B., Foretz M. AMPK Re-activation suppresses hepatic steatosis but its downregulation does not promote fatty liver development. EBioMedicine. 2018;28:194–209. doi: 10.1016/j.ebiom.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Herzig S., Shaw R.J. AMPK: guardian of metabolism and mitochondrial homeostasis. Nature Reviews Molecular Cell Biology. 2017;19:121–135. doi: 10.1038/nrm.2017.95. 2017 19:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hardie D.G., Schaffer B.E., Brunet A. AMPK: an energy-sensing pathway with multiple inputs and outputs. Trends in Cell Biology. 2016;26:190–201. doi: 10.1016/j.tcb.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sakamuri S.S.V.P., Sperling J.A., Sure V.N., Dholakia M.H., Peterson N.R., Rutkai I., et al. Measurement of respiratory function in isolated cardiac mitochondria using Seahorse XFe24 Analyzer: applications for aging research. Geroscience. 2018;40:347–356. doi: 10.1007/s11357-018-0021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]