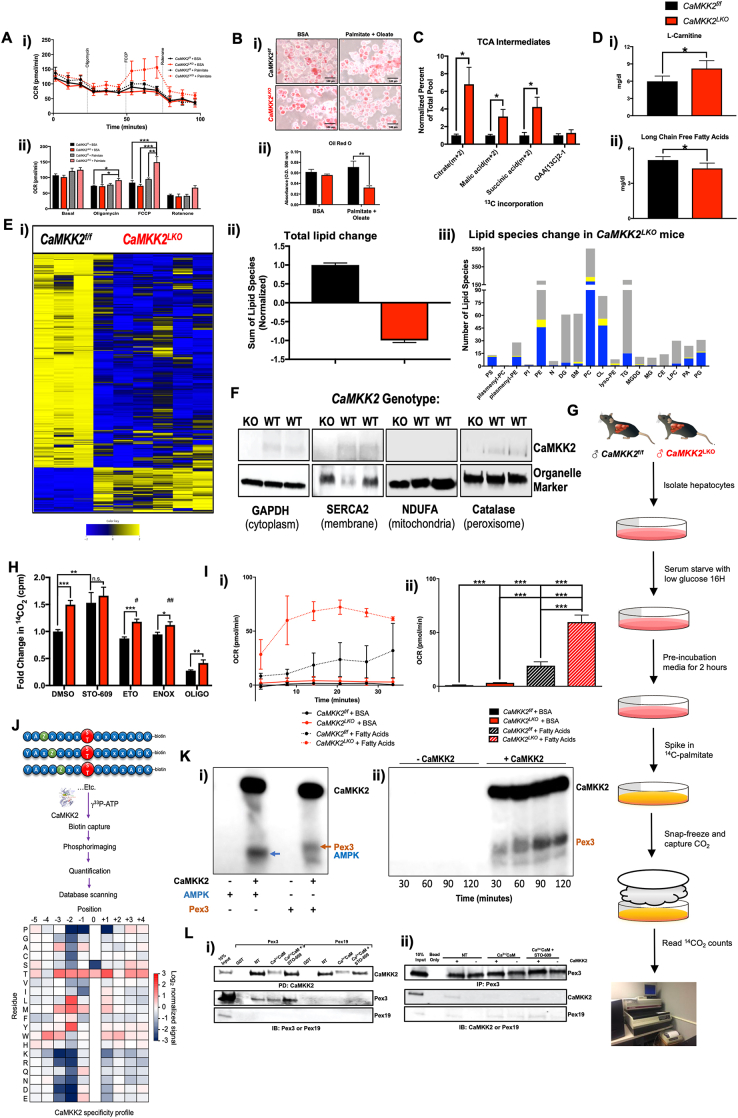
Figure 4.
Hepatic CaMKK2 ablation increases cell autonomous fatty acid metabolism. (A) (i) OCR of primary hepatocytes isolated from CaMKK2f/f (black lines) and CaMKK2LKO (red lines) mice in the presence (dotted lines) or absence (solid lines) of palmitate as measured by Seahorse technology and (ii) average at each stage represented by bar graphs. (B) (i) Primary hepatocytes from (A) were stained with Oil Red O following the Seahorse assay, photographed (ii) and the dye extracted, and absorbance measured at O.D. 500 nm. (C) Primary hepatocytes isolated from CaMKK2f/f and CaMKK2LKO mice were serum starved, followed by addition of FBS and 13C-palmitate or unlabeled palmitate (control) and incubated for 6 h 13C incorporation into TCA cycle intermediates in CaMKK2f/f (black bars) and CaMKK2LKO (red bars) was measured and represented as percent of the total pool normalized to CaMKK2f/f incorporation. (D) Livers from CaMKK2f/f (black bars) and CaMKK2LKO (red bars) were assayed for levels of (i)l-carnitine and (ii) long chain free fatty acids. (E) Lipidomic analysis of livers from CaMKK2f/f and CaMKK2LKO mice: (i) Heatmap representation of the change in lipid species and (ii) quantification of total lipid landscape in livers from CaMKK2f/f (black bars) and CaMKK2LKO (red bars) mice. (iii) Bar graph representing the number and direction of lipid species changes in CaMKK2LKO relative to CaMKK2f/f livers. Blue denotes decreased lipid, yellow denotes increased lipid, and grey denotes lipids that remained unchanged. PE phamenyl-PC (plasmenyl-glycerophophocholines); plasmenyl-PE (plasmenyl-gycerophophoethanolamines); PI (glycerophosphoinositols); PE (gylcerophosphoethanolamines); N (sphinganines); DG (diacylgycerolipids); SM (sphingomyelins); PC (glycerophosphocholines); CL (cardiolipins); lyso-PE (lyso-glycerophosphoethanolamines); TG (triacylgycerolipids); MGDG (monoacylgycerolipids/diacylgycerolipids); MG (monoacylgycerolipids); CE (cholesterol esters); LPC (lyso-glycerophophocholines); PA (glycerophosphates); PG (glycerophosphogycerols). (F) Immunoblot analysis of hepatic CaMKK2 protein expression in the cytoplasmic, membrane, mitochondrial, and peroxisomal fractions. (G) Schematic of β-oxidation assay from primary hepatocytes isolated from CaMKK2f/f and CaMKK2LKO mice, serum starved O/N, incubated in pre-incubation media, and treated with 14C-palmitate. (H) Results from the β-oxidation represented as fold change in 14CO2 counts between CaMKK2f/f (black bars) and CaMKK2LKO (red bars) mice after treatment with DMSO, 20 mM STO-609 (CaMKK2 inhibitor), 20 mM Etomoxir (ETO; mitochondrial Cpt1 inhibitor), 150 mM enoximone (ENOX; peroxisomal phosphodiesterase inhibitor), or 10 mM oligomycin (OLIGO; electron transport chain inhibitor, negative control). #: denotes statistical significance compared to DMSO-treated primary hepatocytes from CaMKK2LKO mice. (I) Peroxisomes isolated from CaMKK2f/f and CaMKK2LKO livers were assayed by Seahorse: (i) OCR of peroxisomes isolated from CaMKK2f/f (black lines) and CaMKK2LKO (red lines) mice in the presence of BSA (solid lines) or fatty acids (dotted lines). (ii) Average of averages of OCR readings from (i). (J) Schematic of degenerate peptide library analysis used to identify putative CaMKK2 phospho-targets and the actual residues identified. (K) (i)In vitro kinase assay with recombinantly purified CaMKK2 and Pex3. AMPK1-312; positive control. (ii) Time-course in vitro kinase assay comparing phosphorylation levels of Pex3 at different timepoints in the absence (lanes 1–4) or presence (lanes 5–8) of CaMKK2. (L) (i) Immunoblot analysis following pull-down of GST-CaMKK2 in the presence of Pex3 or Pex19 under control conditions (NT), activated conditions (Ca2+/CaM), and inhibited conditions (Ca2+/CaM + STO-609). (ii) Immunoblot analysis of CaMKK2, Pex3 and Pex19 following immunoprecipitation of Pex3. Data are represented as the mean ± s.e.m. Unpaired Student's t-test was used to determine statistical significance. One-way ANOVA was used to analyze peroxisomal respiration ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.