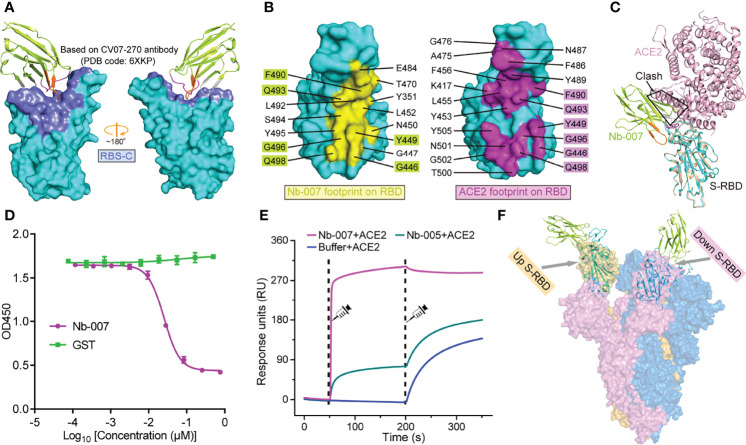
Figure 4.
Molecular basis for Nb-007 neutralization. (A) The previously identified RBS-C epitope [Based on CV07-270 antibody (PDB code: 6XKP)] is colored by slate and mapped onto our Nb-007/S-RBD complex. The relative orientation of RBD is the same as in Supplementary Figure 7. Nb-007 is shown in cartoon representation and colored as in Figure 3. (B) The footprints of Nb-007 (left panel, highlighted in yellow) and ACE2 (right panel, highlighted in magenta) on S-RBD. The involved residues are marked and the overlapping amino acids are highlighted. (C) Superimposition of the complex structures of Nb-007/S-RBD and ACE2/S-RBD. Steric clashes between Nb-007 and ACE2 are highlighted. (D) Competitive binding assays by ELISA. SARS-CoV-2 S-RBD was coated on 96-well plates, recombinant Fc-fused ACE2 was first added, followed by serial dilutions of Nb-007. Error bar stands for the mean ± SD. Experiments were performed in triplicate. (E) SPR kinetics of competitive binding of Nb-007 and ACE2 to SARS-CoV-2 S-RBD. S-RBD was immobilized onto a sensor chip. The indicated nanobodies (Nb-007 and Nb-005) and ACE2 were successively injected. The real-time binding profiles are recorded. Clearly shown is that Nb-007 but not Nb-005 interferes with ACE2 binding. (F) Alignment of the Nb-007/S-RBD structure (shown in cartoon) onto a previously solved cryo-EM structure of the SARS-CoV-2 S-trimer (shown in surface, PDB code: 6VYB). The up- and down-conformation of the S-RBD are highlighted.