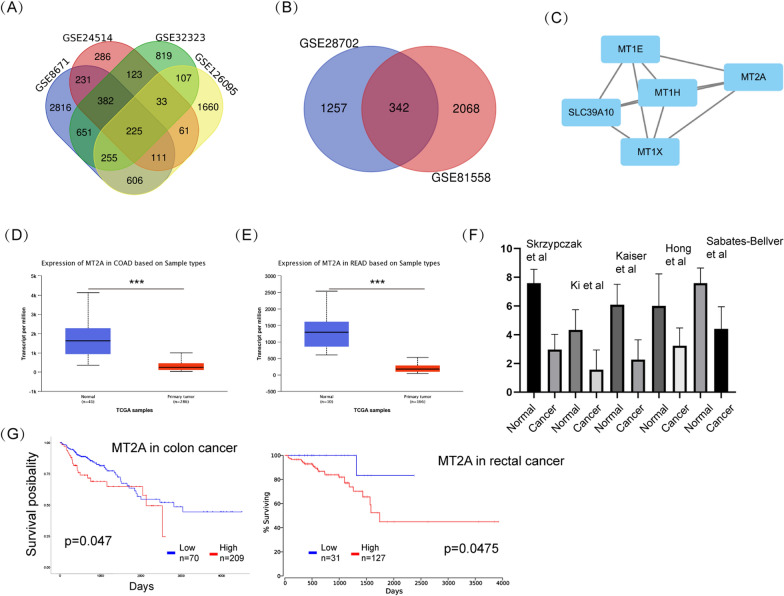
Fig. 1.
MT2A is downregulated in CRC samples. A Four GSE datasets (GSE8671, GSE24514, GSE32323 and GSE126095), which involved data of primary colorectal cancer and paracancerous tissues, were utilized. With a cutoff of p < 0.01 and |log2fold change|> 1, 225 genes were found to be commonly altered in the four datasets. B Two GSE datasets (GSE28702 and GSE81558), were used to compare the gene expression between primary and liver metastasis tumors, and 342 genes were found to be commonly altered. C The MCODE plug-in APP of Cytoscape was used to select the significant module in the PPI network and generate the most likely potential function cluster. Five genes, namely, MT1E, MT2A, MT1H, MT1X and SLC39A10, were included in the predicted cluster. D TCGA data from COAD were used to examine the expression of MT2A in colon cancer, and MT2A was significantly downregulated in cancer tissues compared to normal tissues. E TCGA data from READ were used to examine the expression of MT2A in rectal cancer, and MT2A was significantly downregulated in cancer tissues compared to normal tissues. F Five Oncomine datasets from Skrzypczak et al., Ki et al., Kaiser et al., Hong et al. and Sabates-Bellver et al. were selected, and all showed that MT2A was downregulated in cancer tissues. G Clinical data of patients with colon and rectal cancer were obtained from TCGA, and higher expression of MT2A indicated poor prognosis in both colon and rectal cancer patients. ***p < 0.001