Figure 6.
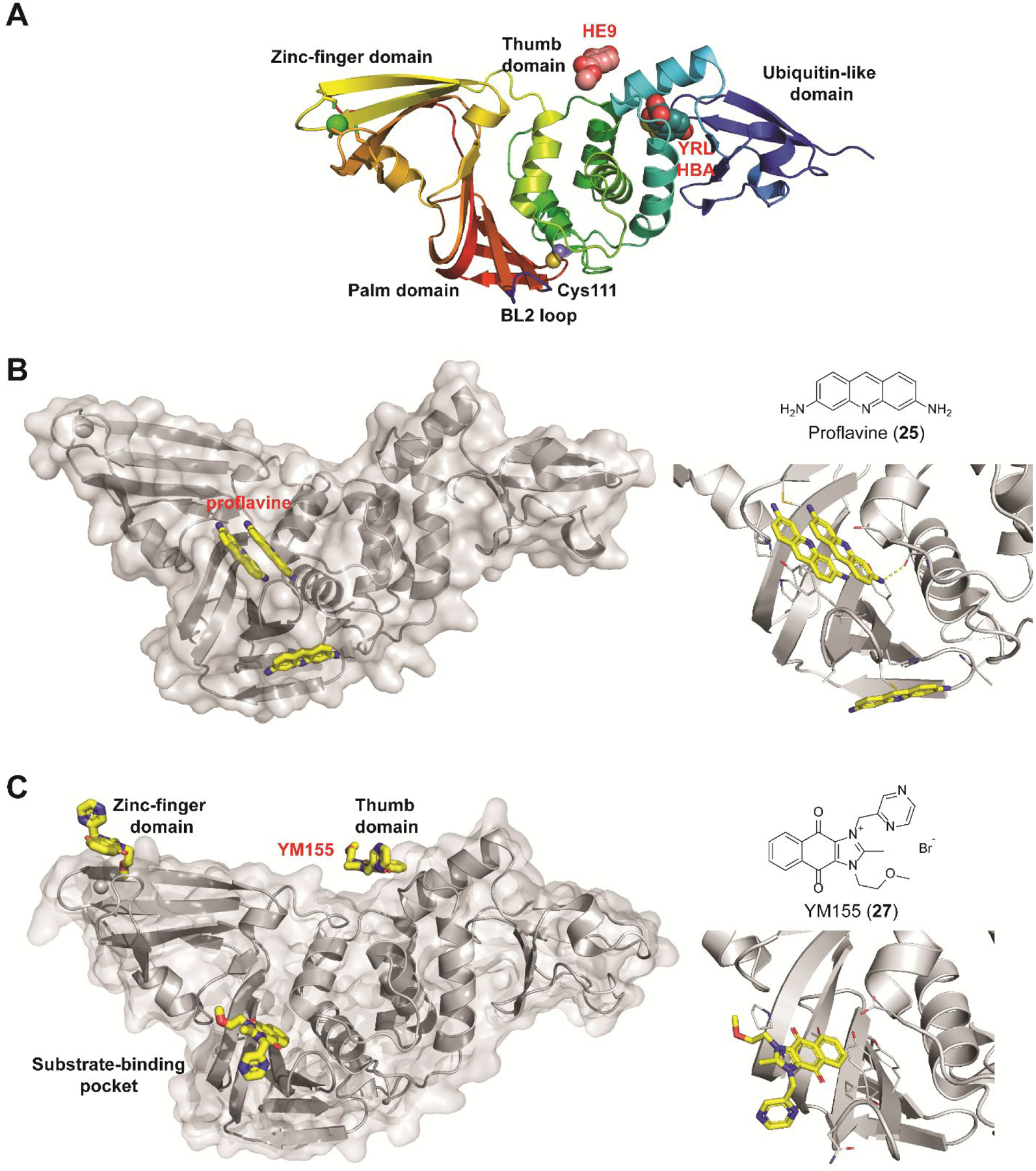
Non-covalent SARS-CoV-2 PLpro inhibitors that do not share structural similarity with GRL0617 (4). (A) X-ray crystal structures of SARS-CoV-2 PLpro in complex with fragments HE9 (20), YRL (21), and HBA (22). (B) X-ray crystal structure of SARS-CoV-2 PLpro in complex with proflavine (25) showing three molecules bind near the BL2 loop (PDB: 7NT4). Two molecules stack on top of each other and fit in the GRL0617 (4) binding pocket, and a third molecule binds at the backside of the BL2 loop. (C) X-ray crystal structures of SARS-CoV-2 PLpro in complex with YM155 (27) (PDB: 7D7L). YM155 (27) binds three different sites located at the zinc-finger domain, thumb domain, and the substrate-binding pocket. Detailed interactions between YM155 and the BL2 loop region residues were shown on the right side.
