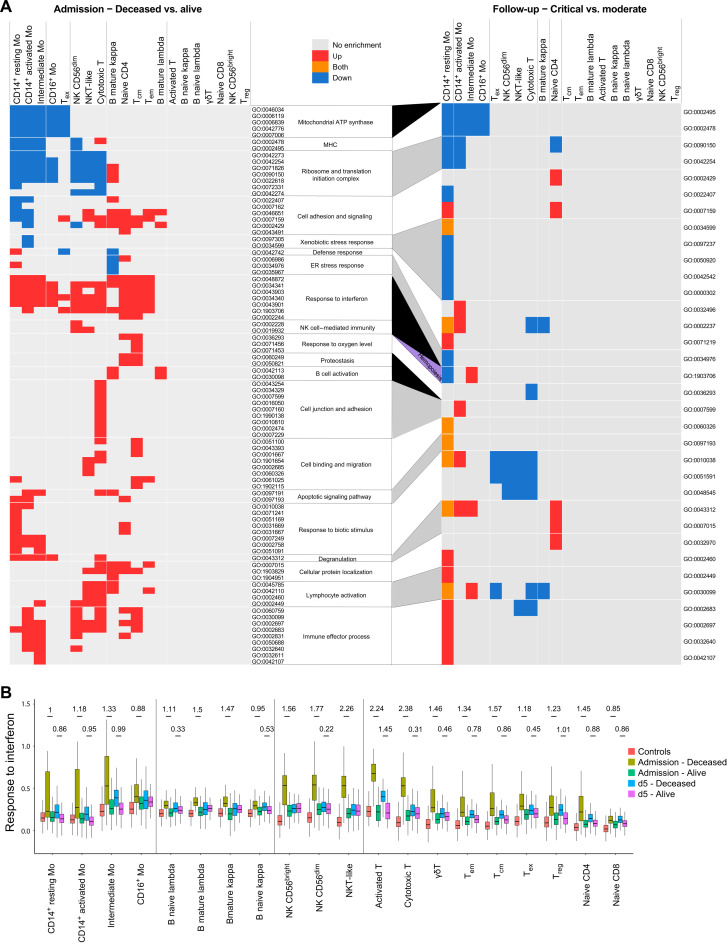
Fig. 3. Transcriptional variations in cell populations of severely ill patients with COVID-19.
(A) GO term enrichment analysis for DEGs at admission between the deceased versus alive patient groups (left heatmap) and between the critical versus moderate patients with COVID-19 at follow-up (right heatmap). Heatmap boxes are colored on the basis of the enrichment: gray for no significant enrichment; blue, red, and orange for significant (q value ≤ 0.05) enrichment of down-regulated, up-regulated, and both up- and down-regulated genes, respectively. GO categories only enriched at admission are highlighted in black between the two heatmaps, whereas the GO category highlighted in yellow (hemopoiesis) represents the only category enriched only at follow-up. GO categories enriched at both admission and follow-up are successively highlighted in white and gray for easier visualization. (B) Evolution of the transcriptional expression for the “response to interferon” GO term. DEGs enriched in the GO term in (A) were used to generate a module score representing the overall expression level for term genes in the cell lineage indicated at the bottom of the graph. Module scores shown were derived at admission and at the day 5 time points for the deceased and alive patients with COVID-19 separately. Control samples from healthy participants are included as reference. Significance of differences between deceased and alive patients with COVID-19 was tested by comparing the corresponding module score distribution in every cell subset using a Wilcoxon rank test approach (q value ≤ 0.05). To provide a sample-size free evaluation of the difference in expression, Cohen’s d effect size estimation is shown for every significant variation. Transcriptional evolution graphs for the other enriched GO categories are provided in fig. S5. Tem, effector memory T cells; Tex, exhausted T cells; Treg, regulatory T cells; gdT, γδ T cells; B cells, either naive or memory cells expressing either the κ or λ light chain.