Figure 3.
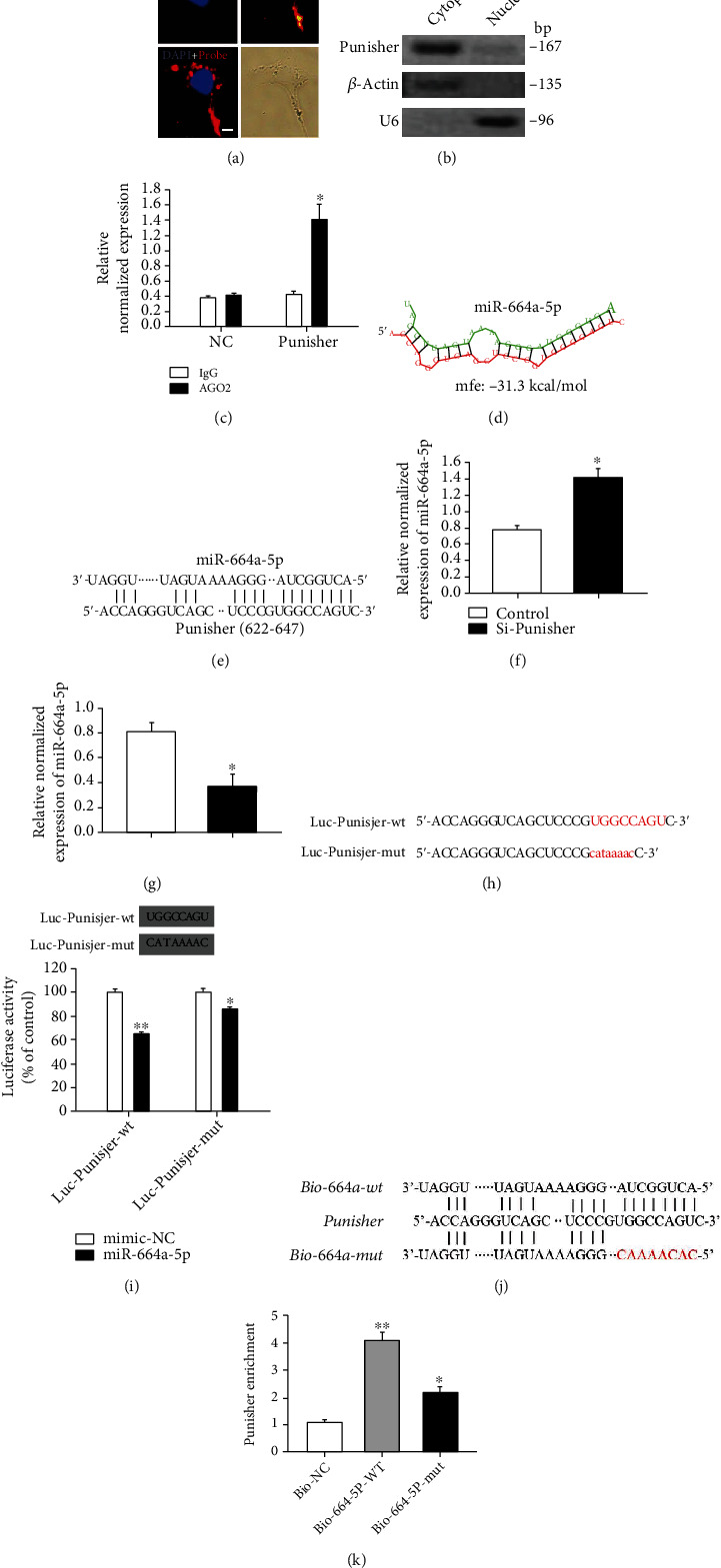
Punisher serves as a miRNA sponge for miR-664a-5p. (a, b) Cellular localization of Punisher in VSMCs by using FISH and western blotting. (c) Assessment of binding potential between Punisher and AGO2 through RIP assay. (d, e) Schematic of the predicted miR-664a-5p binding sites with Punisher. (f, g) qRT-PCR analysis of transcript level of miR-664a-5p after Punisher knockdown or overexpression. (h) The prediction of coding sequence in luciferase constructs with the wild type (Luc-Punisher-wt) and mutated (Luc-Punisher-mut). (i) Luciferase assay of HEK 293 cells cotransfected with a scrambled control, miR-664a-5p mimics, and a luciferase reporter containing Punisher-wt or mutant constructs. (j, k) Pulldown assay with biotinylated miRNA to test the association between Punisher and miR-664a-5p. VSMCs were transfected with biotinylated WT miR-664a-5p (Bio-664-wt) or biotinylated mutant miR-664a-5p with disruption of basepairing between Punisher and miR-664a-5p (Bio-664-mut). A biotinylated miRNA that is not complementary to Punisher was used as a NC (Bio-NC). 24 h after transfection, cells were harvested for biotin-based pulldown assay using streptavidin magnetic beads (Sigma). The bound RNAs were purified using TRIzol for the analysis, and Punisher expression levels were analyzed by qRT-PCR. Data are shown as mean ± s.e.m. of three independent experiments. Scale bars: 20 μm. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
