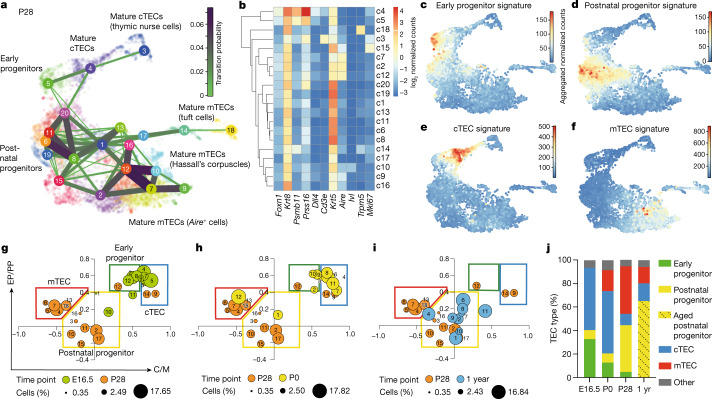
Fig. 1. Heterogeneity of TECs.
a, UMAP representation of transcriptome similarities among 6,959 individual TECs derived from 4-week-old wild-type male (n = 2) and female (n = 2) mice. Cell clusters and transition probabilities were inferred with VarID44; connections with probability P > 0.001 are shown, with transition probabilities indicated by line thickness and colour. The positions of clusters containing early and postnatal bipotent progenitors and mature cTEC and mTEC clusters are indicated. Colours mark cells in the identified cell clusters. b, Expression profiles of signature genes in individual TEC clusters. c–f, UMAP plots highlighting the aggregated expression profiles of gene groups distinguishing early (c) and postnatal (d) progenitors and cTECs (e) and mTECs (f). g–i, Age-dependent changes in the TEC compartment. Transcriptome features of TEC clusters are shown at various time points expressed as ratios of progenitor and mature TEC gene set transcript counts; the P28 time point was used as a reference. Assignment of clusters to the four main populations in the coordinate system is indicated in g; the sizes of dots correspond to the relative fraction in the TEC population. j, Summary of dynamic changes in the composition of the TEC compartment. yr, year.