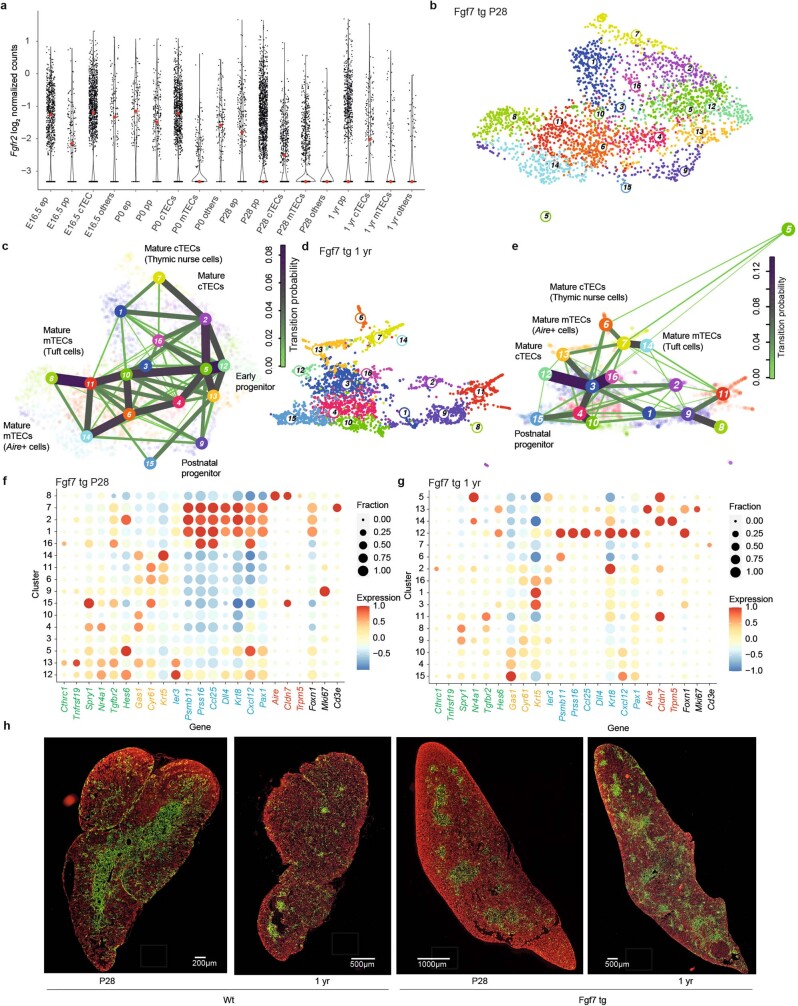
Extended Data Fig. 13. Effect of continuous Fgf7 signalling on the TEC compartment in young and old mice.
a, Expression of the Fgfr2 gene in cells of different TEC subsets of mice at different time points (E16.5 EP, n = 721 cells; E16.5 PP, n = 166 cells; E16.5 cTECs, n = 1,159 cells; E16.5 others, n = 141 cells; P0 EP, n = 136 cells; P0 PP, n = 226 cells; P0 cTECs, n = 924 cells; P0 mTECs, n = 311 cells; P0 others, n = 148 cells; P28 EP, n = 292 cells; P28 PP, n = 2,302 cells; P28 cTECs, n = 554 cells; P28 mTECs, n = 2,338 cells; P28 others, n = 303 cells; 1yr PP, n = 981 cells; 1yr cTECs, n = 225 cells; 1yr mTECs, n = 209 cells; 1 yr others, n = 87 cells). Data are presented as violin plots; the red dots indicate median expression levels. Negative cells are given a pseudo-count of 0.1. scRNA-seq datasets of barcoding mice across different time points were merged and normalized by downscaling to 1,500 transcript counts in order to calculate the log2-normalized transcript counts for Fgfr2. b, d, UMAP representation of transcriptome similarities between individual TECs isolated from thymi of P28 (b), or 1 year-old (d) Foxn1:Fgf7 transgenic mice. c, e, Cluster designations deduced by VarID indicating the transcriptional relationships in terms of VarID-derived transition probabilities; connections with probabilities P > 0.001 are shown and the transition probabilities are indicated by line thickness and colouring. For orientation purposes, the major cell populations are also indicated. In panel (e), cluster 5 represents cells derived from ectopic parathyroid tissue. f, g, Expression profiles of TEC clusters for the indicated signature genes and the two time points. The fractions of each cluster expressing a particular gene and their respective expression levels are depicted according to the scales shown on the right. Dot colour represents the z-score of the mean expression levels of the gene in the respective cluster and dot size represents the fraction of cells in the cluster expressing the gene; gene names are coloured according to shared expression patterns (EP: green; PP: orange; cTEC, blue; mTEC, red; other genes of interest, black). z-scores above 1 and below -1 are replaced by 1 and -1 respectively. h, Immunohistochemical analysis of thymic lobes of wild-type (wt) and Foxn1:Fgf7 transgenic mice at two different time points. Sections were stained with anti-Keratin 5 (green) and anti-Keratin 18 (red) antibodies, marking medullary and cortical compartments. Scale bars are indicated; panels are representative of 4 mice.