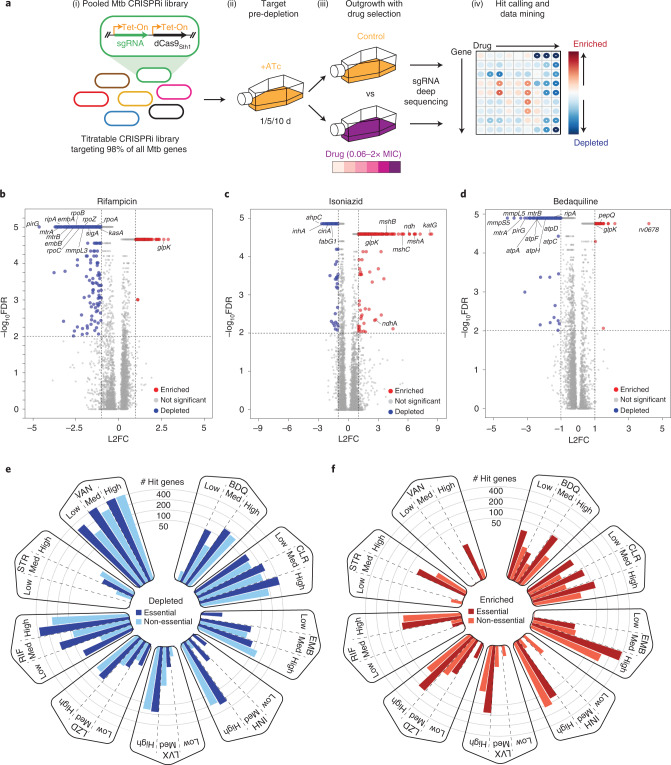
Fig. 1. Chemical-genetic profiling identifies hundreds of genes that influence drug efficacy in M. tuberculosis.
a, Quantifying chemical-genetic interactions in Mtb. (i) The pooled CRISPRi library contains 96,700 sgRNAs targeting 4,052/4,125 Mtb genes. In vitro essential genes15 were targeted for titratable knockdown by varying the targeted PAM and sgRNA targeting sequence length; non-essential genes were targeted only with the strong sgRNAs. (ii) The CRISPRi inducer ATc was added for 1, 5 or 10 d before drug exposure to pre-deplete target gene products. (iii) Triplicate cultures were outgrown +ATc in DMSO or drug at concentrations spanning the predicted MIC. (iv) Following outgrowth, genomic DNA was collected from cultures treated with three descending doses of partially inhibitory drug concentrations (‘High’, ‘Med’ and ‘Low’; Extended Data Fig. 1), sgRNAs amplified for deep sequencing, and hit genes called with MAGeCK. Growth phenotypes were highly correlated among triplicate screens (average Pearson correlation between replicate screens: r > 0.99). b–d, Volcano plots showing log2 fold change (L2FC) values and false discovery rates (FDR) for each gene for the indicated drugs (‘High’ concentration, 1 d CRISPRi library pre-depletion for RIF and 5 d for INH and BDQ). e,f, The number of significantly depleted and enriched hit genes (FDR < 0.01, |L2FC| > 1) are shown for the indicated drugs. Hit genes were defined as the union of 1 and 5 d target pre-depletion screens because these datasets recovered the majority (>95%) of unique hits (Extended Data Fig. 2). Gene essentiality was defined by CRISPRi15.