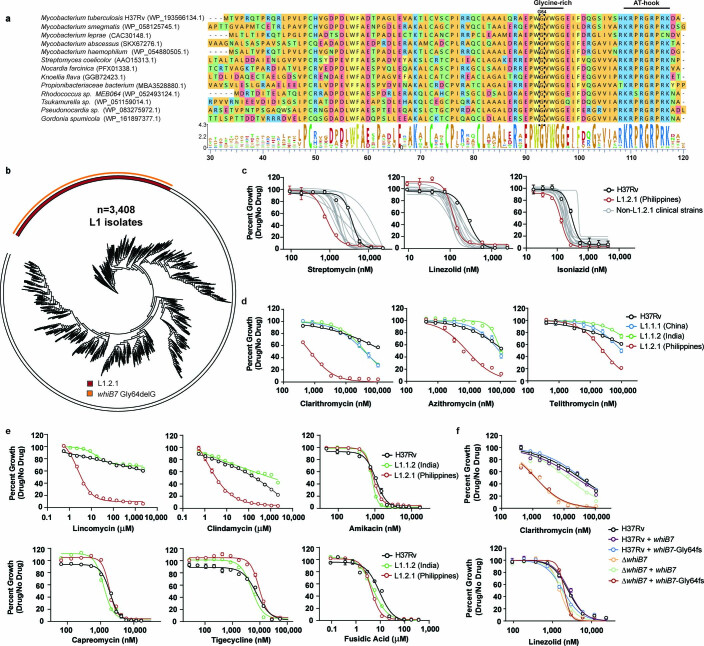
Extended Data Fig. 8. L1.2.1 is a whiB7 loss-of-function mutant and hypersusceptible to macrolides, ketolides, and lincosamides.
a, Alignment of WhiB7 orthologues from representative actinobacteria. Accession numbers are listed next to each species. The conserved glycine-rich motif and DNA binding AT-hook element are highlighted. b, Phylogenetic tree of all L1 Mtb clinical isolates (n = 3,408) in our genome database (Source Data Fig. 4). L1.2.1 and the whiB7 Gly64delG mutation are highlighted. c-e, Dose-response curves (mean ± SEM, n = 3 biological replicates) for a reference set of Mtb clinical strains66. f, H37Rv or a ΔwhiB7 H37Rv strain were transformed with an integrating plasmid to express either the H37Rv whiB7 allele or the L1.2.1 whiB7 Gly64delG allele in trans from its native promoter. Dose-response curves (mean ± SEM, n = 3 biological replicates) for the resulting strains are shown.