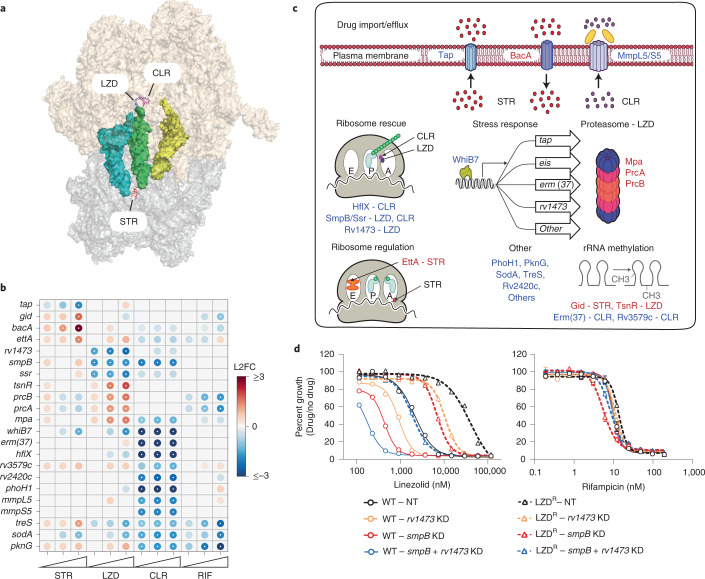
Fig. 3. Diverse pathways contribute to intrinsic resistance and susceptibility to ribosome-targeting antibiotics.
a, Structure of LZD, CLR and STR bound to the Thermus thermophilus ribosome. PDB codes: 3DLL, 1J5A, 1FJG, 4V5C. b, Heatmap depicting chemical-genetic interactions as in Fig. 2a. c, Chemical-genetic hit genes from Fig. 3b are involved in diverse cellular pathways. Genes whose inhibition decreased or increased fitness in the indicated drug are listed in blue or red, respectively. d, Dose-response curves (mean ± s.e.m., n = 3 biological replicates) for the indicated CRISPRi strains in H37Rv or rplC-Cys154Arg linezolid-resistant H37Rv (LZDR).