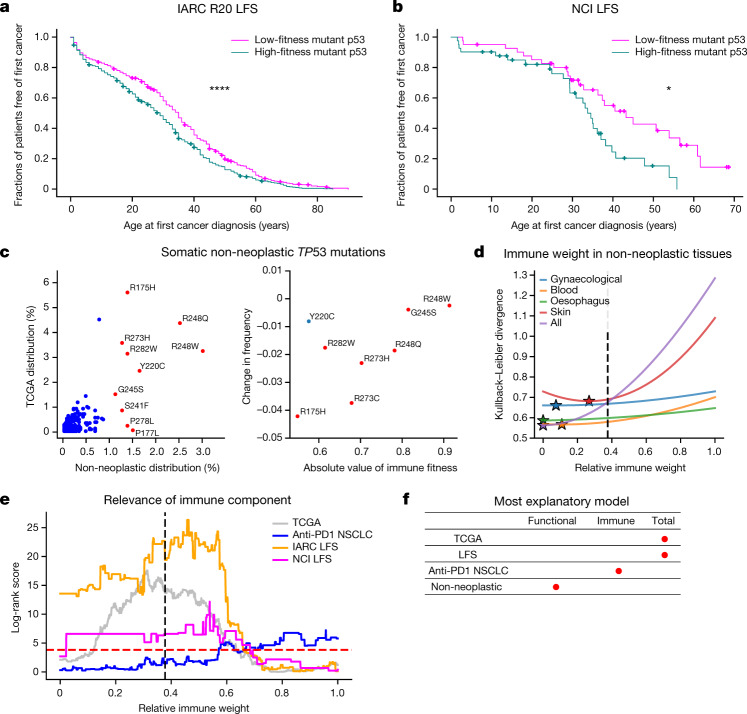
Fig. 4. Mutant p53 fitness informs LFS age of tumour onset and non-neoplastic TP53 mutation distribution.
a, b, Kaplan–Meier curves split on median mutant p53 fitness from the combined model for age of tumour onset in the IARC R20 germline dataset (n = 998) (a) and the NCI LFS dataset (n = 82) (b). c, Left, comparison of TP53 mutation frequencies in non-neoplastic tissues (3,451 mutation occurrences) and the frequencies in TCGA (2,764 mutation occurrences; Pearson r = 0.732, P < 0.0001; Spearman r = 0.544, P < 0.0001; top 10 non-neoplastic mutations coloured in red and annotated). Right, positive relationship between hotspot frequency difference in non-cancerous and cancerous cells and magnitude of immune fitness. CpG-associated hotspots are coloured in red; Y220C is coloured in blue (overall: Pearson r = 0.594, P = 0.120; Spearman r = 0.619, P = 0.102; CpG-associated hotspots only: Pearson r = 0.827, P = 0.022; Spearman r = 0.786, P = 0.036). d, Kullback–Leibler divergence plotted as a function of relative immune weight for the largest tissue-specific mutation distributions across collected non-neoplastic somatic p53 mutations. Optimal immune weights are denoted as stars, and the optimal relative immune weight derived independently to best represent the observed mutation frequency in TCGA is denoted as a black dotted line. e, Log-rank scores of the TCGA (n = 1,941), NSCLC (n = 289) and LFS (IARC, n = 946; NCI, n = 82) cohorts as a function of the relative immune weight. The dashed red line corresponds to the log-rank score for P = 0.05; the dashed black line marks the choice of parameters trained independently to best represent the observed mutation frequency in TCGA. f, The most explanatory models across mutant TP53 datasets, as indicated by red dots.