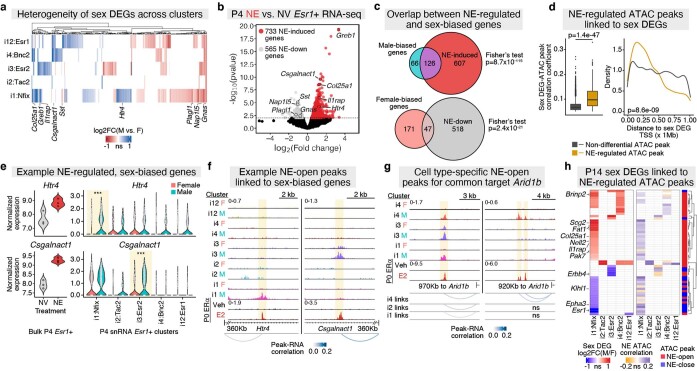
Extended Data Fig. 11. Sex differences in single-nucleus multiome dataset.
a, Hierarchical clustering of log2FC values for P4 sex DEGs detected in Esr1+ inhibitory neuron clusters (see also Fig. 3e). Sex DEGs with non-significant differential expression colored in white. b, Neonatal E2 (NE) vs. neonatal vehicle (NV) female nuclear RNA-seq on P4 BNST Esr1+ cells; grey, red dots (DESeq2, padj < 0.1). c, (top) Overlap between NE-induced genes and P4 multiome male-biased genes. (bottom) Overlap between NE-downregulated genes and female-biased genes. d, (left) Pearson’s correlation coefficient values for non-differential (grey) and NE-regulated (gold) ATAC peaks that correlate with P4 sex DEG expression. Boxplot center=median, box boundaries=1st and 3rd quartile, whiskers=1.5*IQR from boundaries. n = 5169 non-differential, 244 NE-regulated. p-value from two-sided, Wilcoxon rank-sum test. (right) Distance between non-differential (grey) and NE-regulated (gold) ATAC peaks to P4 sex DEG transcription start sites (TSS). p-value from Kolmogorov-Smirnov test. e, Example P4 sex-biased genes that are also NE-regulated, Htr4 (top) and Csgalnact1 (bottom). (left) n = 3, (right) n = 887 i1:Nfix female cells, 676 i1:Nfix male cells, 404 i3:Esr2 female cells, 550 i3:Esr2 male cells. f, Tracks for NE-open ATAC peaks that correlate with NE-regulated, sex-biased targets, Htr4 and Csgalnact1. g, Different NE-open ATAC peaks across i1:Nfix, i3:Esr2, and i4:Bnc2 neurons correlated with a common male-biased target, Arid1b. h, Heatmaps indicating (left) RNA log2FC of P14 sex DEGs and (right) Pearson’s correlation coefficient of NE-open (red) and -close (blue) ATAC peaks linked to sex DEGs within each cluster. Non-significant genes and correlation values colored in white.