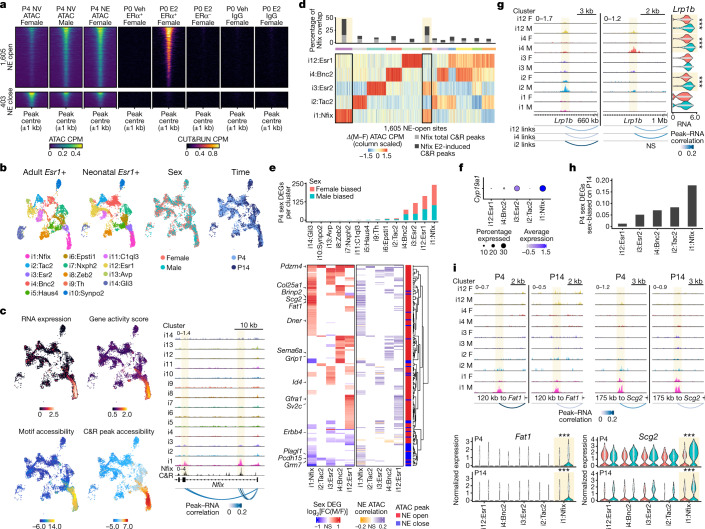
Fig. 3. Neonatal ERα genomic binding drives a sustained male-biased gene expression program.
a, Heatmap of P4 BNST Esr1+ ATAC, P0 IgG CUT&RUN and P0 ERα CUT&RUN CPM ±1 kb around 1,605 NE-open and 403 NE-close ATAC peaks (edgeR, Padj < 0.1). ERα+, Sun1–GFP+ nuclei; ERα−, Sun1–GFP− nuclei. b, UMAPs of adult (left) and neonatal (middle left) BNST Esr1+ snRNA-seq clusters; neonatal snRNA-seq clusters coloured by sex (middle right) and time point (right). c, Left: UMAPs of Nfix expression (top left), gene activity score (top right), motif chromVAR deviation score (bottom left) and CUT&RUN chromVAR deviation score (bottom right). Right: neonatal single-nucleus ATAC (snATAC) and adult BNSTp Nfix CUT&RUN tracks at the Nfix locus. Top left number is the y-axis range in CPM. Shaded band indicates peak region. Peak–RNA correlation indicates correlation coefficient for snATAC peaks correlated with Nfix expression. d, Heatmap of differential snATAC CPM between males (M) and females (F) at 1,605 NE-open sites, scaled across snRNA-seq clusters and grouped using k-means clustering. The barplot indicates the percentage of overlap for each k-means cluster with total and E2-induced BNSTp Nfix CUT&RUN peaks. e, Top: number of sex DEGs (MAST, Padj < 0.05) in P4 multiome clusters. Bottom: heatmaps indicating RNA log2[FC] of P4 sex DEGs (left) and Pearson’s correlation coefficient of NE-open (red) and NE-close (blue) ATAC peaks (right) linked to sex DEGs in each cluster. Genes without significant differential expression or correlation coefficients (not significant (NS)) are shown in white. f, Cyp19a1/aromatase expression on P4. g, Left: NE-open ATAC peaks correlating with Lrp1b expression in Cyp19a1− clusters, i2:Tac2 and i12:Esr1. Top left number is the y-axis range in CPM. Shaded band indicates peak region. Right, sex difference in Lrp1b expression in i2:Tac2 (n = 260 female, 153 male, Padj = 2.13 × 10−8), i4:Bnc2 (n = 437 female, 373 male, Padj = 5.62 × 10−37), i12:Esr1 (n = 803 female, 507 male, Padj = 1.09 × 10−12) cells. ***Padj < 0.001, MAST. h, Proportion of P4 sex DEGs detected as sex biased on P14. i, Top: i1:Nfix-specific, NE-open ATAC peaks at Fat1 and Scg2 loci on P4 and P14. Top left number is the y-axis range in CPM. Shaded band indicates peak region. Bottom: Sex difference in i1:Nfix Fat1 and Scg2 expression on P4 (Fat1, Padj = 1.28 × 10−37; Scg2, Padj = 1.54 × 10−46; n = 887 female, 676 male) and P14 (Fat1, Padj = 1.13 × 10−11; Scg2, Padj = 1.52 × 10−5; n = 554 female, 829 male). ***Padj < 0.001, MAST.