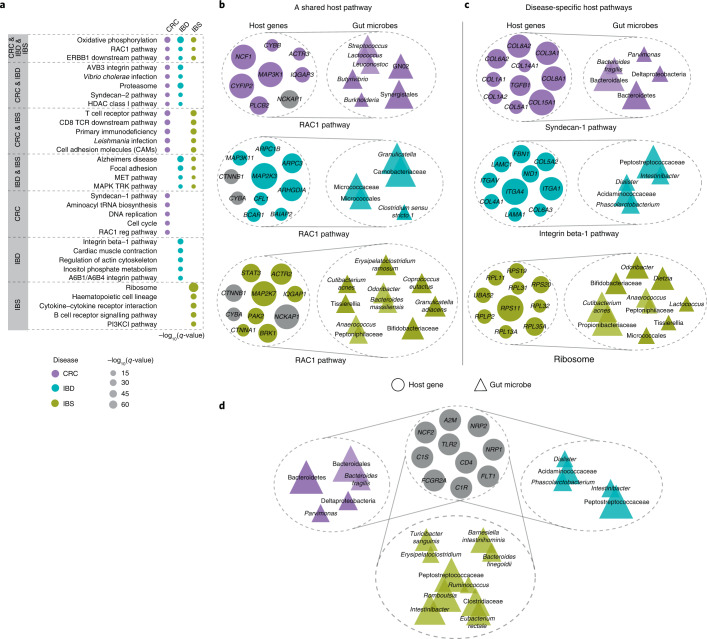
Fig. 2. Shared immunoregulatory and metabolic host pathways associate with disease-specific gut microbes across human diseases.
a, Host pathways enriched for sparse CCA gene sets associated with gut microbiome composition across diseases (FDR < 0.1). Dot size represents the significance of enrichment for each pathway, and dot colour denotes the disease cohort in which this pathway is significantly associated with microbiome composition. b, Association between microbial taxa in CRC, IBD and IBS (top to bottom) and host genes in the RAC1 pathway, a pathway for which host gene expression correlates with gut microbes across disease cohorts (a shared host pathway). The size of circles and triangles represents the absolute value of sparse CCA coefficients of genes and microbes, respectively. Microbial taxa belonging to a common taxonomic order are shown as overlapping triangles. Genes that are common between pathways or components across at least two disease cohorts are shown in grey. c, Association between the set of host genes in disease-specific host pathways (that is, host pathways for which gene expression correlates with gut microbes in only one of the disease cohorts) and groups of gut bacteria in CRC, IBD and IBS (top to bottom). d, A common set of host genes (grey circles) that are associated with disease-specific sets of microbes. These host genes are enriched for immunoregulatory and acute inflammatory response pathways.