Fig. 5. Cryo-EM maps of a ≈ 5 MDa 60-subunit Cage-GIDSR4 assembly.
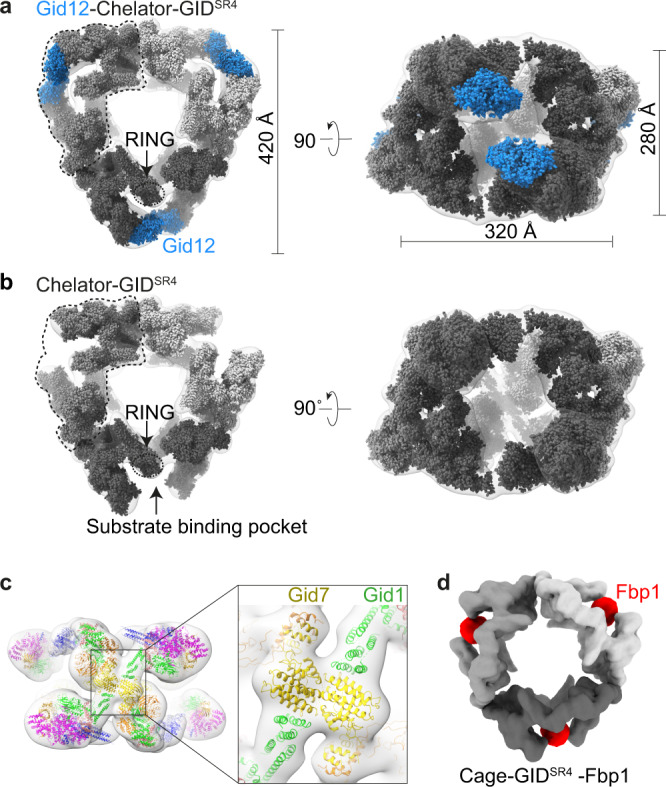
a Overall architecture of Gid12-Cage-GIDSR4 is seen from its transparent cryo-EM density fit with three copies of models of Gid12-Chelator-GIDSR4 in solid surfaces in different shades of gray, with models for Gid12 (this study) in blue. b 19.8 Å-resolution cryo-EM density for Cage-GIDSR4 is shown as a transparent surface fit with three copies of models of Chelator-GIDSR4 (EMD-12541) in solid surfaces in different shades of gray. c Models of Gid1 and Gid7 domains fitted at junctions of the three individual Chelator-GIDs. These interaction at the junction constitute the cage architecture. The resolution of the density, together with prior maps and models, allows attributing domains but not their specific orientations or interactions. d 12 Å-resolution cryo-EM density map of Cage-GIDSR4-Fbp1 (here, the N-terminal degron of Fbp1 - PTLVNG was exchanged with Mdh2 degron - PHSVTP to increase stability). Fbp1 tetramer encapsulated in the oval center of each Chelator-GIDSR4 is color-coded in red.
