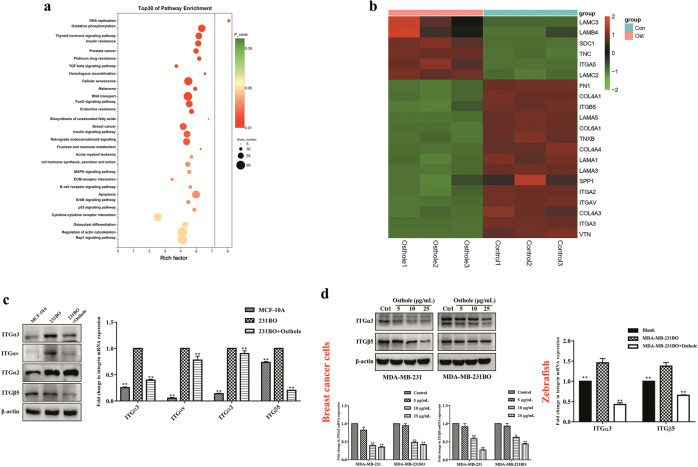
Fig. 3. Identification of candidate target genes of osthole in highly bone-metastatic breast cancer cells by RNA sequencing.
a KEGG pathways enrichment analysis of differentially expressed genes (DEGs) in MDA-MB-231BO cells after osthole treatment. b Heatmap of DEGs between MDA-MB-231BO cells and MDA-MB-231BO + osthole cells (|log2 Fold Change| > 1, P < 0.05). c Identification of the candidate osthole-targeted genes using Western blot and real-time PCR. **P < 0.01 vs. the MDA-MB-231BO group. d Regulatory effect of osthole on ITGα3 and ITGβ5 in highly metastatic breast cancer cells (MDA-MB-231 and MDA-MB-231BO) and the zebrafish breast cancer xenograft model induced by MDA-MB-231BO cells. Data were expressed as the mean ± SD for triplicate. For breast cancer cells, *P < 0.05, **P < 0.01 vs. the control group; for the zebrafish model, **P < 0.01 vs. the MDA-MB-231BO group.