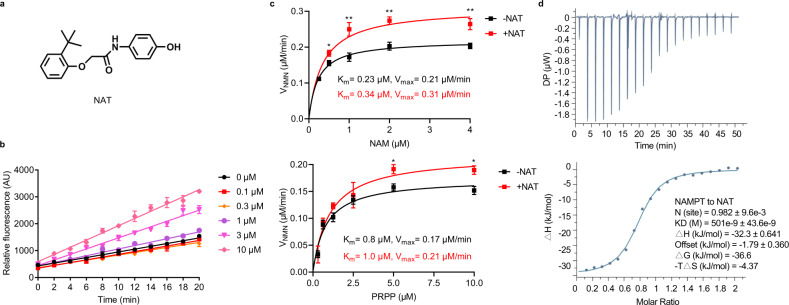
Fig. 1. NAT binds and directly activates NAMPT.
a Chemical structure of NAT. b Dose-dependent activation of NAMPT by NAT. A triply-coupled enzyme assay was performed to monitor the conversion of NAM to NADH. The first enzyme, NAMPT, converted NAM to NMN. The second enzyme, NMNAT1, converted NMN to NAD. The third enzyme, ADH, converted NAD to NADH. The reactions were carried out at room temperature for 20 min in the presence of increasing concentrations of NAT. Fluorescence of NADH was measured at Ex340/Em445 (y-axis). Progress curves of the enzyme reactions were plotted and data were analyzed with GraphPad Prism. Data are represented as means ± SEM from three replicates in a representative experiment. n = 3 independent experiments. c Effect of NAT on the NAMPT enzyme kinetics in the direct NAMPT assay. Michaelis–Menten curves for NAMPT reactions were plotted in the presence or absence of 10 μM NAT. The velocity of NMN production is shown on the y-axis as a function of substrate concentration (x-axis). Upper panel: Michaelis–Menten curves to determine the kinetic parameters for substrate NAM. Reactions contained the indicated concentrations of NAM and 10 μM PRPP. Lower panel: Michaelis–Menten curves to determine the kinetic parameters for substrate PRPP. Reactions contained the indicated concentrations of PRPP and 5 μM NAM. Vmax, maximum reaction rate; Km, substrate affinity. All error bars represent SEM from three replicates. Two-tailed t-test, *P < 0.05, **P < 0.01. d Binding of NAT to recombinant NAMPT as measured by isothermal titration calorimetry (ITC). A total of 200 μM recombinant NAMPT was titrated into the sample cell containing 20 μM NAT. The data shown here is a representative figure from three independent experiments. Top panel, the integrated heat signatures. Bottom panel, the fitted curves using one-site mode. KD, the dissociation constant; ΔG, change in Gibbs energy of binding; ΔH enthalpy; -TΔS, the entropy contribution to Gibbs energy; N, stoichiometry. Data were analyzed with MicroCal PEAQ-ITC Analysis software.