Figure 2.
Functional genomic screening of the GRACE PK library identifies regulators of antifungal drug tolerance
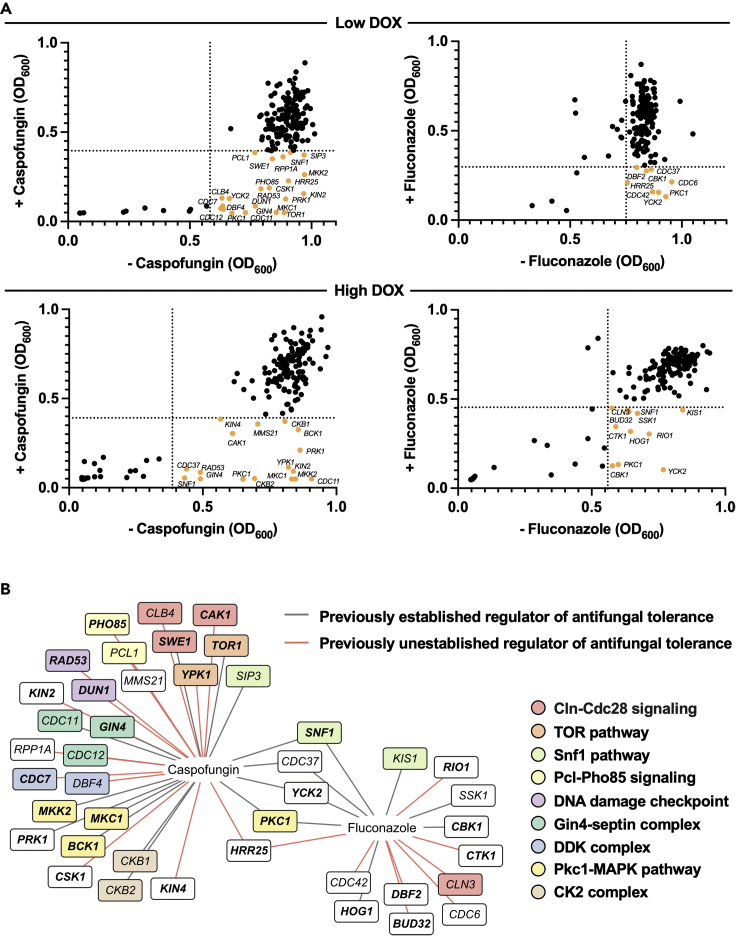
(A) Scatter (XY) plots depicting the growth of each strain in the presence and absence of caspofungin or fluconazole upon DOX-mediated transcriptional repression. The GRACE PK library was grown overnight with low DOX (0.05 μg/mL) or high DOX (20 μg/mL). Cells were subsequently inoculated into YPD with the corresponding DOX, in the presence and absence of caspofungin (15 ng/mL) or fluconazole (0.8 μg/mL). Growth (OD600) was measured after 24 and 48 h of incubation at 30°C for the caspofungin and fluconazole screens, respectively. Data points are averages of technical duplicates. Dotted lines indicate the median absolute deviation (MAD) cut off values for each condition. Hits are labeled and displayed in orange. See also Figure S1.
(B) Hits identified from the antifungal drug susceptibility screens. Previously established and unestablished regulators of antifungal tolerance are distinguished by gray and red lines, respectively. Functional groups are indicated by colored boxes (see legend). PKs are specified in bold text. PK regulators are unbolded.