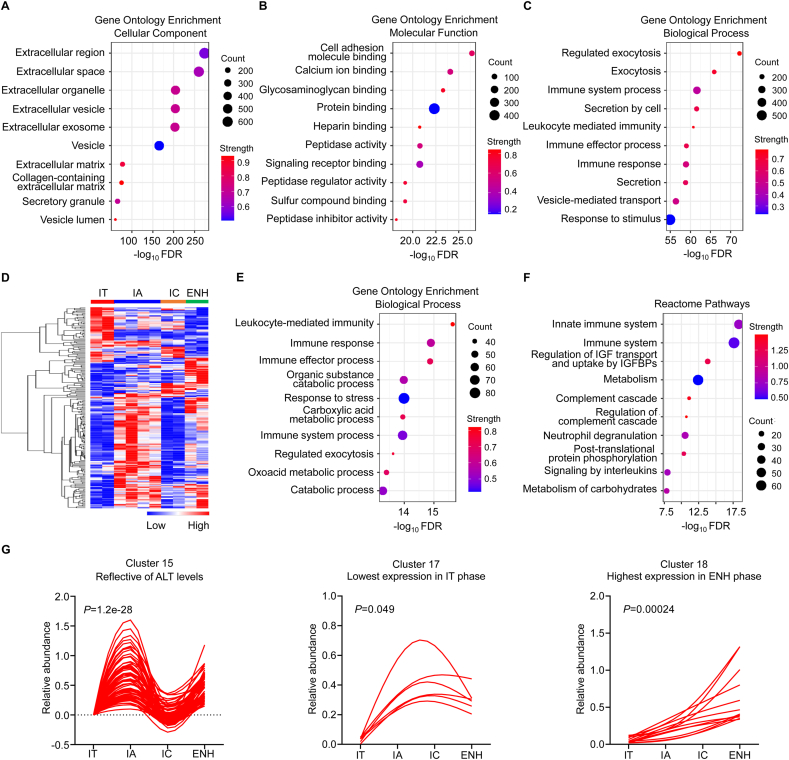
Fig. 2.
Serum proteomic characterization of the natural history of chronic HBV infection.(A–C) Gene Ontology (GO) enrichment analyses of the identified 1162 proteins from pooled serum were done to evaluate enriched cellular components, molecular functions, and biological processes among the natural history of chronic HBV infection. (D) Heatmap of serum proteomic profiles in different disease phases. (E and F) GO Terms and Reactome pathway enrichment analyses of 226 differentially expressed proteins were performed to evaluate enriched biological processes and signaling pathways in different phases of chronic HBV infection. (G) Serum protein pattern analyses by the Short Time-series Expression Miner (STEM). FDR, false discovery rate; IT, immune-tolerant phase; IA, immune reactive HBeAg-positive phase; IC, inactive HBV carrier state phase; ENH, HBeAg-negative chronic hepatitis B phase.