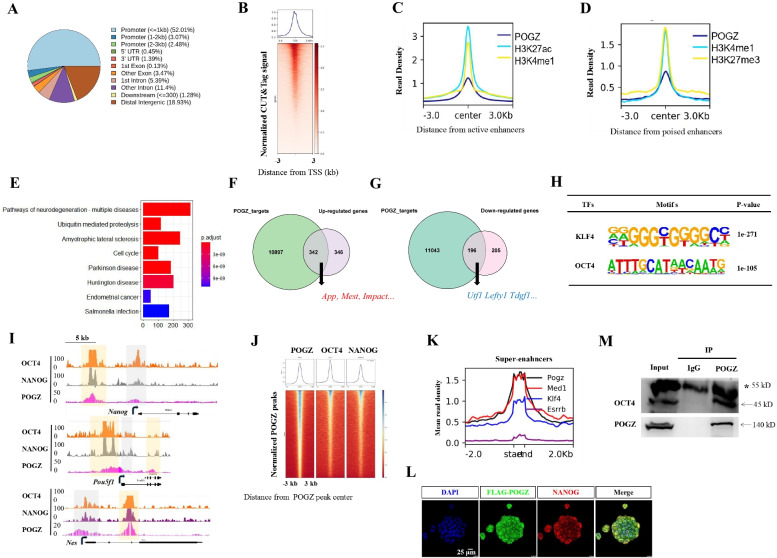
Fig. 4.
POGZ directly activates and represses target genes. A Pie chart showing the distribution of POGZ binding sites genome wide. B Heat map of POGZ CUT&Tag enrichment in a 6 kb window around the TSS. C Metaplot of POGZ, H3K4me1 and H3K27ac enrichment (normalized per million mapped reads) on ± 3 kb of genes that were bound by POGZ (27,343 peaks). D Metaplot of POGZ, H3K4me1 and H3K27me3 enrichment (normalized per million mapped reads) on ± 3 kb of genes that were bound by POGZ (1808 peaks). E KEGG analysis of POGZ-bound genes in ESCs. F Pie chart showing the overlap of POGZ targets and up-regulated genes. G Overlap of POGZ targets and down-regulated genes. H The enrichment of KLFs and OCT4/SOX2 binding motifs at POGZ-bound loci, as revealed by HOMER. I CUT&Tag and ChIP-seq snapshots showing the co-localization of POGZ, NANOG and OCT4 at the indicated loci. Gray: proximal TSS; Orange: distal regions. J Heat map of NANOG, OCT4 and POGZ signals across sites bound by POGZ (n = 11,000). Each row represents a 6 kb window centered on the peak midpoint. K Metaplot of POGZ, MED1, ESRRB and KLF4 enrichment (normalized per million mapped reads) on ± 2 kb window across all 231 super-enhancers that have been identified in ESCs by Whyte [41]. L Double IF staining of NANOG and FLAG-POGZ in ESCs. Bar: 25 μm. M IP results showing that OCT4 interacts with POGZ in ESCs. Star pointing to the IgG heavy chain. All IF and WB experiments were repeated at least two times