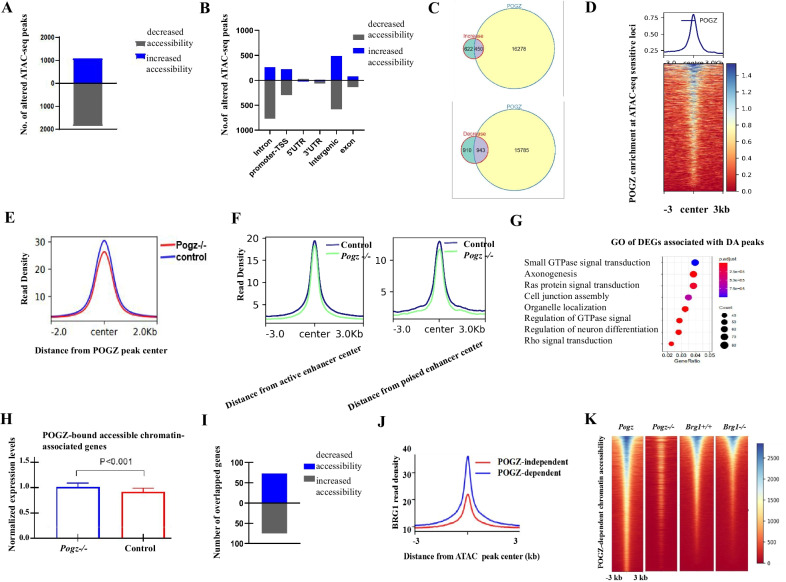
Fig. 6.
POGZ regulated-chromatin accessibility is linked with BRG1. A Bar chart showing number of altered ATAC-seq peaks. Blue color: ATAC-seq peaks showing increased signals in Pogz−/− ESCs; Black color: ATAC-seq peaks showing decreased signals in Pogz−/− ESCs. B Bar chart showing the genome-wide distribution feature of altered ATAC-seq peaks. C Up: Pie chart showing the overlap of POGZ peaks and ATAC-seq peaks showing increased signals in Pogz−/− ESCs; Bottom: Pie chart showing the overlap of POGZ peaks and ATAC-seq peaks showing decreased signals in Pogz−/− ESCs. D Heat map showing that POGZ signals were enriched at ATAC-seq hypersensitive sites. E Metaplot showing that chromatin accessibility is reduced at all POGZ-bound loci. F Metaplot showing that chromatin accessibility is moderately reduced at both active and poised enhancers, in the absence of POGZ. G GO analysis of DEGs associated with DA peaks. H Bar graph showing that POGZ-bound accessible chromatin-associated genes were slightly up-regulated in Pogz−/− ESCs. I Bar chart showing the overlap of DEGs and genes with altered chromatin accessibility. Seventy-three up-regulated genes were associated with DA regions showing increased chromatin accessibility, and 75 down-regulated genes were associated with DA regions showing decreased chromatin accessibility. J Metaplot showing that BRG1 is highly enriched at POGZ-dependent ATAC sites. K Heat map showing a reduction of ATAC-seq signals at POGZ-dependent ATAC sites in BRG1-depleted ESCs