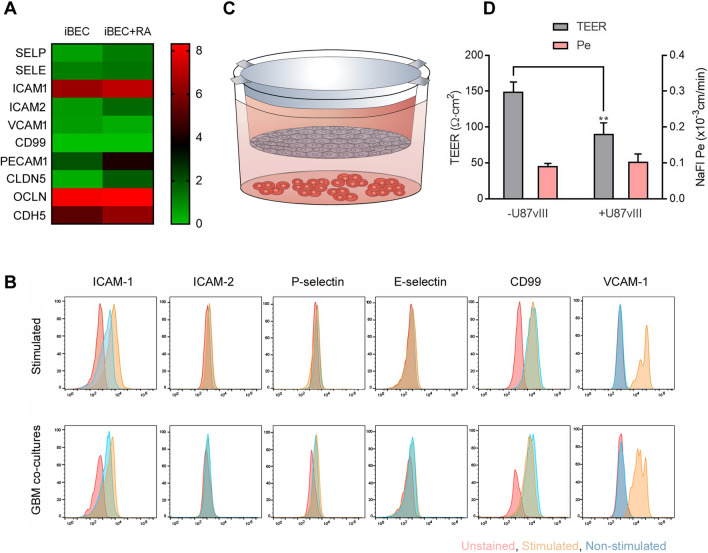
Fig. 1.
Expression of immune cell adhesion molecules on iBECs. A Heatmaps depicting log2 transformed transcript abundances of the immune cell adhesion and BBB-specific genes in iBEC in the absence and presence of Retinoic Acid (+ RA) treatment. Green low-expression, Red high- expression. B Cell surface analysis for expression of adhesion molecules ICAM-1, ICAM-2, P-selectin, S-selectin, CD99 and VCAM1 in iBECs using flow cytometry under naïve (non-stimulated—blue) and cytokine (top panel) and GBM co-culture (bottom panel) stimulated (yellow) conditions. Red—unstained controls. See Additional file 4: Table S2 for MFI values. Representative data shown from 2 independent differentiations. C Schematic illustrating set up of transwell blood–brain barrier (BBB) and U87vIII co-culture model. D Assessment of TEER (Ω cm2) following 24 h co-culture of iBEC transwell inserts with ( +) and without (−) U87vIII cells. TEER and NaFl Pe values are expressed as mean + standard deviation (SD). Statistical significance marked by asterisks assessed by Student T-test where **P ≤ 0.01 (n = 3)