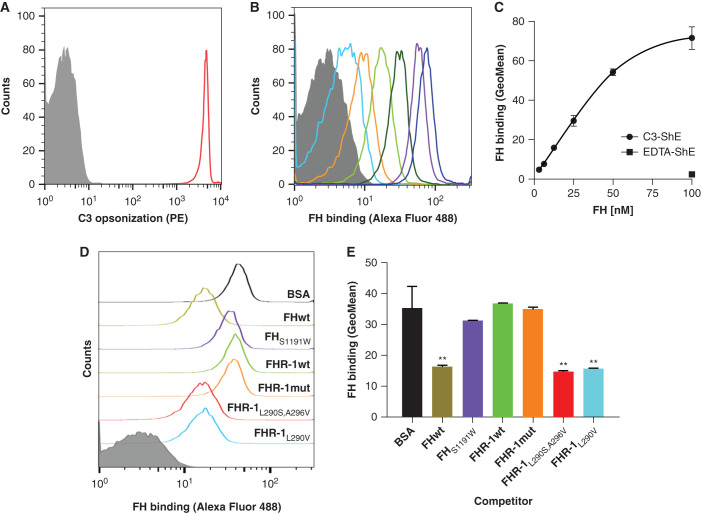
Figure 4.
FH binding to C3b deposited in a physiological surface is not competed by wild-type FHR-1 nor the mutant protein with duplicated dimerization domains. (A) First, antibody-sensitized ShEs were C3b opsonized by incubating cells with NHS depleted of FH and FB (NHSΔFH/FB) in the presence of OmCI to prevent C5 activation. As a negative control, ShEs incubated with NHSΔFH/FB in the presence of EDTA (EDTA-ShE) were used. Opsonization of the cells (C3-ShE, red curve) was confirmed with an in-house monoclonal mouse anti-human C3 antibody (12.17) and a goat anti-mouse IgG phycoerythrin-conjugated antibody (PE) and, as expected, EDTA-ShE (gray curve) were not stained. (B and C) Second, the binding of increasing amounts (3.125–100 nM) of FHAF488 to the ShE was analyzed and showed a FHAF488 dose–dependent binding to the opsonized C3-ShE, whereas the binding to nonopsonized EDTA-ShE was absent at the maximum protein concentration. Competitive binding of 50 nM FHAF488 to C3-ShE was then performed using equimolar amounts of FHwt, FHS1191W (a mutant that does not have the capacity to bind sialic acids), FHR-1wt, FHR-1mut, and the aHUS-associated mutant proteins FHR-1L290S,A296V and FHR-1L290V. Equimolar amounts of BSA were used as a negative control and, hence, represent the binding of FHAF488 to C3-ShE when there is no competition. As can be seen by (D) the flow cytometry curves or (E) by the geometric mean (GeoMean) representations, neither FHR-1wt, FHR-1mut, nor FHS1191W compete with the binding of FHAF488 to cells. Only the aHUS-associated FHR-1 mutant proteins or FHwt significantly displace the FHAF488 from the C3-ShE. Representative experiments of a minimum of two independent experiments are shown in all cases. Data in (E) correspond to the mean±SD of triplicates. **P<0.01, t test.