Figure 3. CRAC/CARC motifs are not predictive of cholesterol binding in human GPCRs.
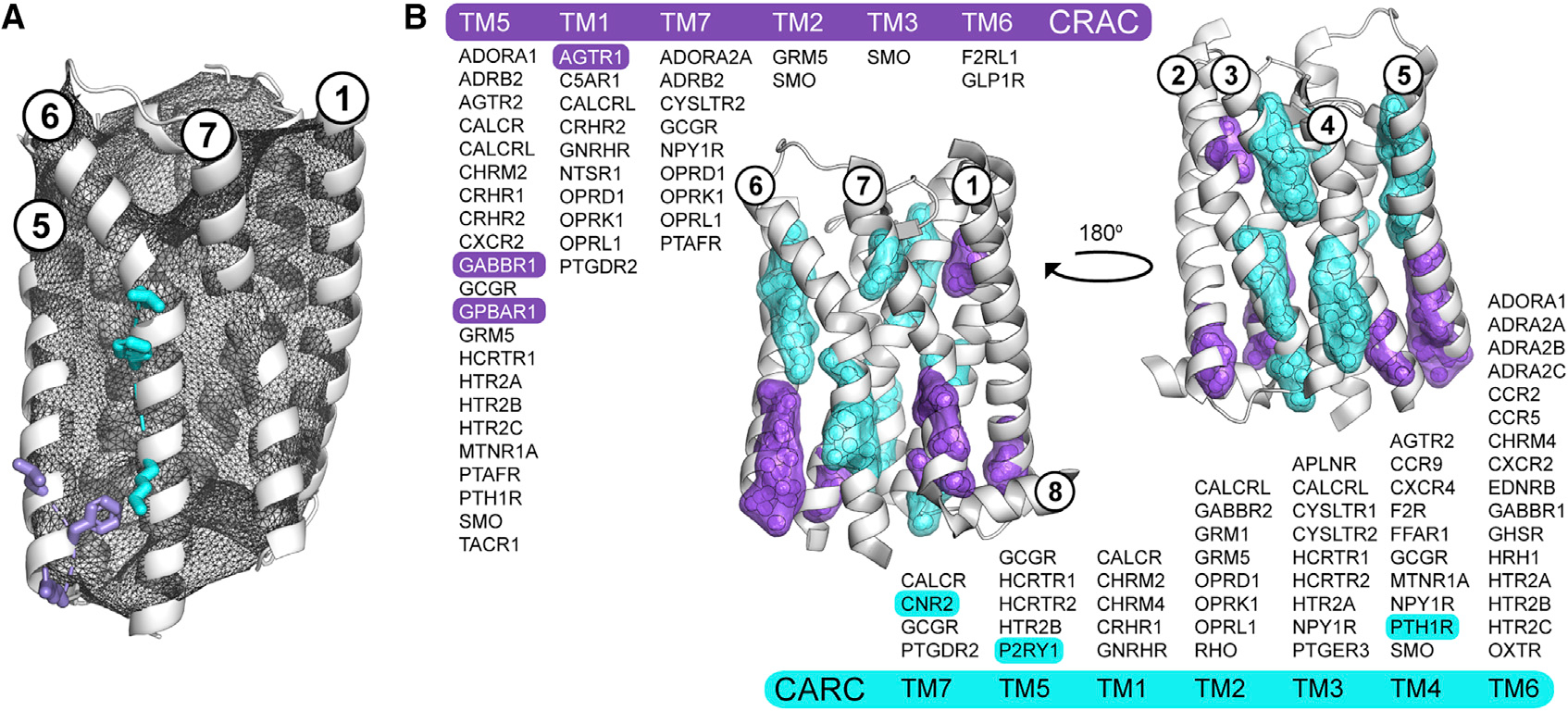
(A and B) Overview of the procedure for validating the location and directionality of CRAC/CARC motifs in GPCRs using the adenosine A1 receptor (ADORA1, PDB code 5N2S, chain A) and two of its putative CRAC (L201, F204, R208; purple) and CARC (K234, F241, L245; cyan) motifs (A). Valid CRAC/CARC motifs met two geometric criteria: the three side chains comprising the motif are located outside the pHinder surface (shown in black mesh) and oriented in the same general direction. Triangulation and clustering of the 164 and 186 putative CRAC (purple) and CARC (cyan) motifs in 473 GPCR structural chains (B). Each motif was first converted to a geometric centroid prior to triangulating and clustering with a minimum cluster size of five. The subset of GPCRs with validated CRAC/CARC motifs are listed by transmembrane helix (TM). Only six of these motifs (highlighted) had CLR observed nearby in a structure. CRAC clusters containing contributions from TM3 (SMO) and TM6 (F2RL1, GLP1R), and CARC clusters containing contributions from TM1 (CACLR, CHRM2, CHRM4, GNRHR) are not shown because they did not meet the minimum cluster size.
