Fig. 9.
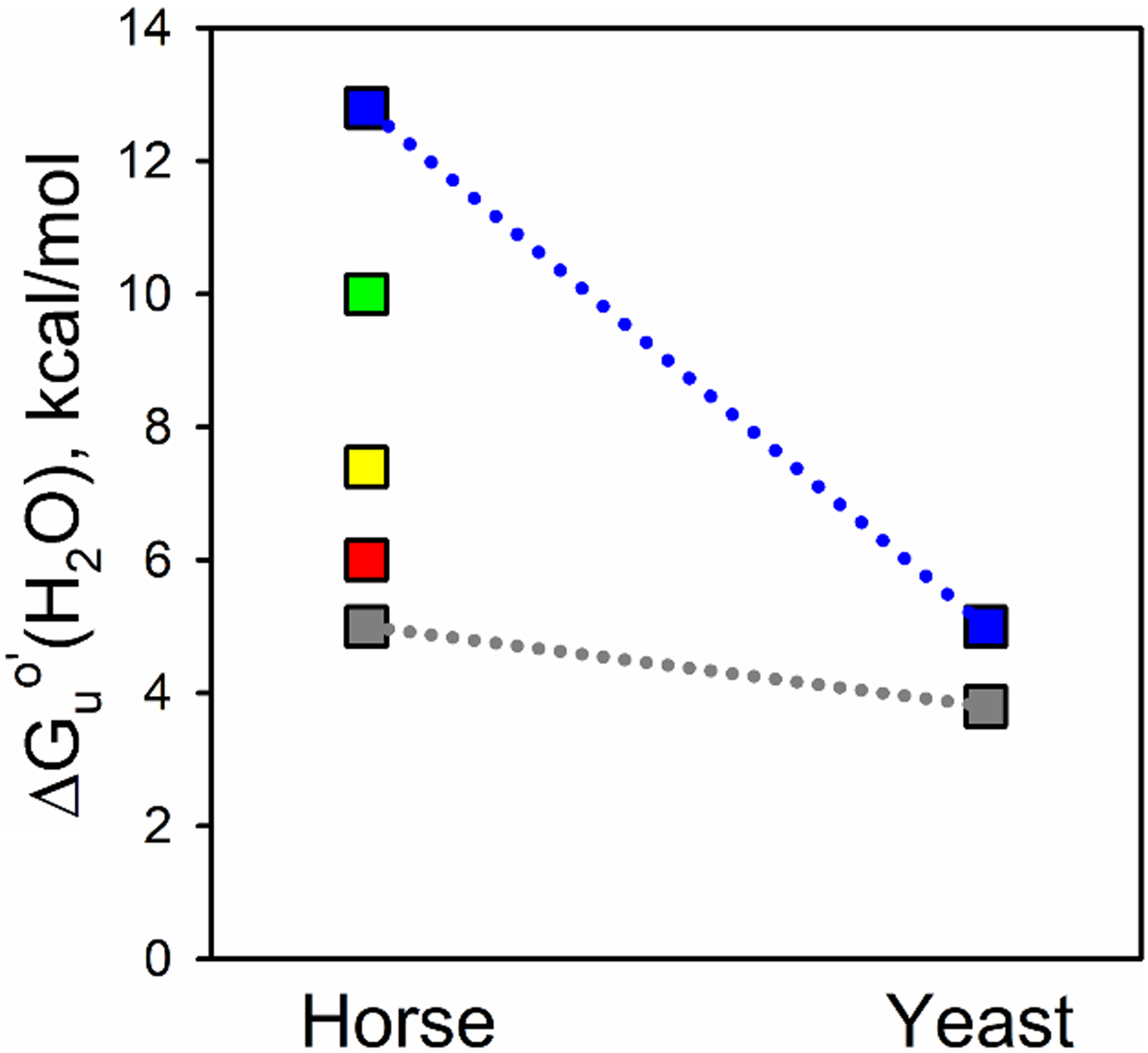
Substructure energy levels for horse Cytc and yeast iso-1-Cytc. The foldons as determined for horse Cytc, using Englander’s nomenclature, are from lowest to highest energy, the infrared (Ω-loop C, shown in gray), red (Ω-loop D), yellow (residues 37 – 39 and 58 – 60), green (residue 19 – 36 Ω-loop and 60s helix), and blue (N- and C-terminal helices) foldons [72]. Limited proteolysis was used to determine the energy of the least stable substructure of yeast iso-1-Cytc [31], which precludes measuring the energies of the other substructures. ΔGuo′(H2O) for unfolding of iso-1-Cytc by GdnHCl was used for the stability of the most stable substructure because the m-value for global unfolding of the yeast protein is similar to that for the most stable structure of horse Cytc obtained from HX experiments.
