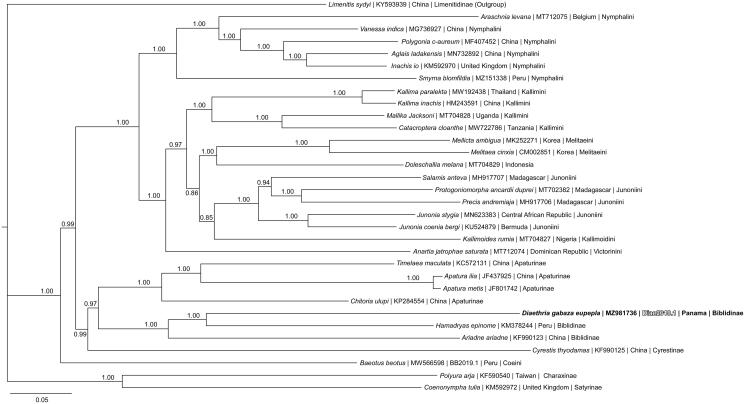
Figure 1.
The Bayesian phylogeny (GTR + I + G model, best state likelihood = −147,240.16, average deviation of split frequencies = 0.001131) of the Diaethria gabaza eupepla mitogenome, 31 additional mitogenomes from within family Nymphalidae, including outgroup species Limenitis sydyi (Nymphalidae: Limenitinae), produced by 10 million MCMC generations in MrBayes, with sampling every 1000 generations, and after discarding the first 250,000 generations as burn-in. The Bayesian posterior probability values determined by Mr Bayes are provided at each node. Each taxon in the analysis is labeled with species name, GenBank accession, the country of origin of the specimen with the sequenced mitogenome, and the nymphalid Tribe or Subfamily of the species.