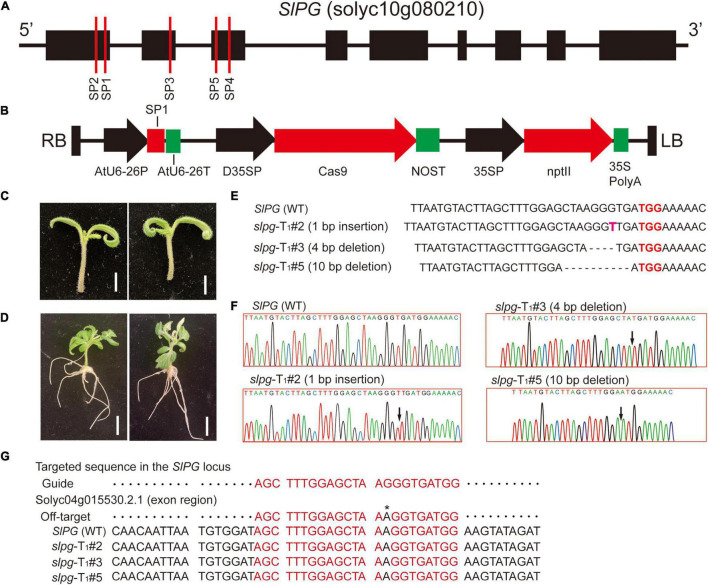
FIGURE 1.
CRISPR/Cas9 technology was used to induce the targeted mutagenesis of the SlPG gene in tomatoes. (A) Schematic representation of tomato SlPG gene and target sites used in this study. Black boxes and lines represent exons and non-coding regions, respectively. Red vertical lines indicate the target site position and name of designed sgRNAs. (B) Schematic illustrating the pg1 vector. RB/LB, right/left border of T-DNA; AtU6-26P, Arabidopsis U6-26 promoter; SP1, sgRNA-SP1; AtU6-26T, Arabidopsis U6-26 terminator; D35SP, double cauliflower mosaic virus (CaMV) 35S promoter; Cas9, coding region of Cas9; NOST, Nos terminator; 35SP, 35S promoter; nptII, kanamycin resistance gene; 35S PolyA, 35S PolyA terminator. (C) Tomato seedlings were selected as explants used for transient transformation. Scale bar, 1 cm. (D) Well-developed hairy roots. Scale bar, 1 cm. (E) Sequences of WT plant and T1 homozygous slpg mutants (slpg-T1#2, slpg-T1#3, and slpg-T1#5) and representative mutation types induced at target site SP1 are presented, respectively. Nucleotides in red represent PAM sequences. Nucleotide in pink represents insertions and dashes between nucleotides represent deletions. (F) Sequence peaks of WT plant and T1 homozygous slpg mutants (slpg-T1#2, slpg-T1#3, and slpg-T1#5). The black arrowheads indicate the location of mutations. (G) Representative putative off-target site recognized by the gRNA designed in this study. One representative site was predicted by the website tool CRISOR. The sequence at the top of the alignment is predicted off-targeted sequence. Red sequences, nucleotides corresponding to the site recognized by the gRNA designed in this study; Black nucleotide with * on top, nucleotide mismatch in the sgRNA region.