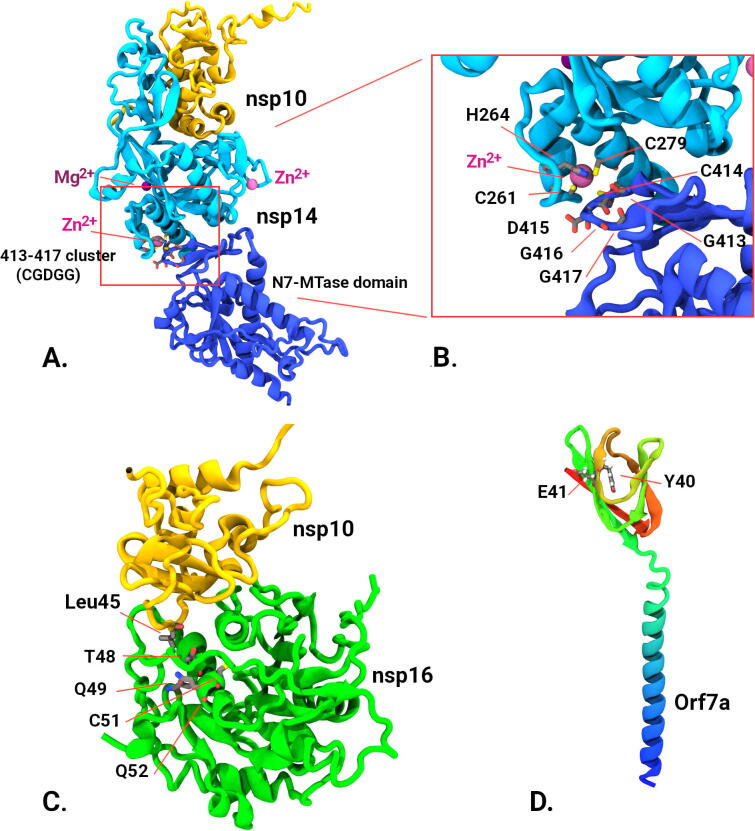
Fig. 3.
Structural analysis of protein models. Alphafold models (verified with crystallographic data where possible). A) 3′-5′ exonuclease (nsp14) (cyan and blue) complexed with nsp10 (yellow) showing relative positions of 413:417 cluster in the N7-MTase domain to Zn binding residues and other ion binding sites. B) Close up of the 413–417 cluster in nsp14 showing proximity of Zn binding domain. C) Structure of nsp10/nsp16 complex (from pdb; 6W4H and Alphafold models) showing nsp16 mutational cluster (T48, Q49, C51, Q52) and its proximity to nsp10 binding, in particular with residue Leu45 form nsp10. D) Predicted Alphafold model of Orf7a accessory protein showing putative mutation sites Y40 and E41. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)