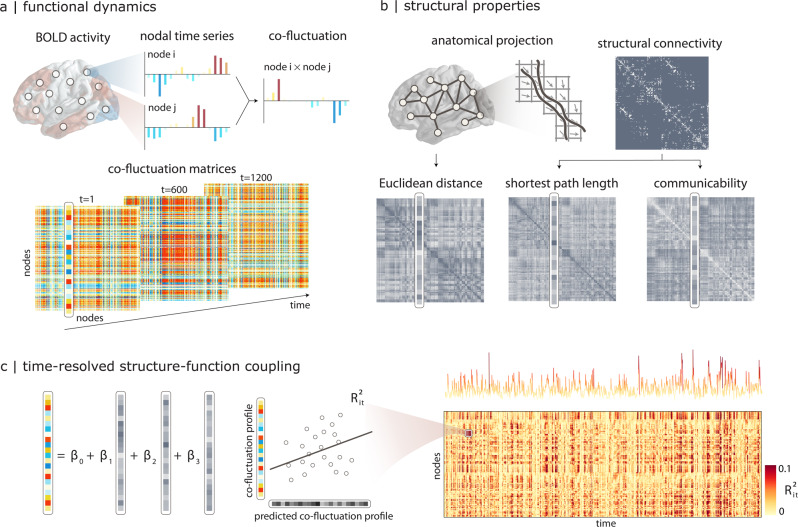
Fig. 1. Time-resolved structure-function coupling.
a The co-fluctuation of two brain regions i and j is calculated as the element-wise multiplication of the two z-scored fMRI BOLD activity time-series. The points of this time-series can be represented as one element in a co-fluctuation matrix. b Pairwise structural relationships are derived from structural connectivity networks reconstructed from diffusion MRI, including Euclidean distance between node centroids, shortest path length and communicability. c A multilinear regression model is used to predict a region’s co-fluctuation profile from its structural profile, using Euclidean distance, path length and communicability as predictors. The resulting coefficient of determination () indicates how well the structural connectivity profile predicts the functional connectivity profile for a particular brain region i at a particular time point t. The procedure generates a region × time matrix that captures the fluctuation of structure-function coupling for individual regions across time. The time-series shows time-resolved fluctuations in mean R2.